4R1T
 
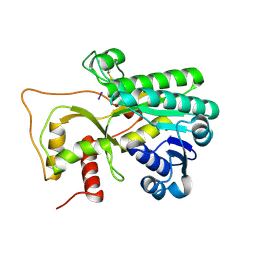 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | Descriptor: | cinnamoyl CoA reductase, molecular iodine | | Authors: | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
1KWI
 
 | | Crystal Structure Analysis of the Cathelicidin Motif of Protegrins | | Descriptor: | Protegrin-3 Precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
3J0F
 
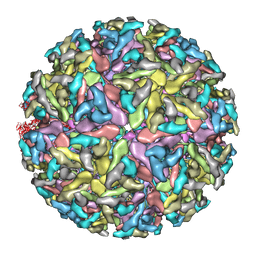 | | Sindbis virion | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Tang, J, Jose, J, Zhang, W, Chipman, P, Kuhn, R.J, Baker, T.S. | | Deposit date: | 2011-07-08 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Molecular Links between the E2 Envelope Glycoprotein and Nucleocapsid Core in Sindbis Virus.
J.Mol.Biol., 414, 2011
|
|
6M6Y
 
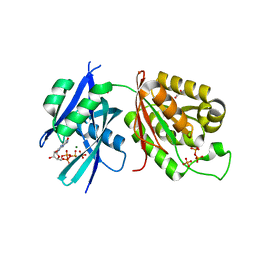 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZUK
 
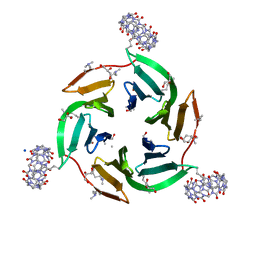 | | Crystal structure of dimethylated RSL-N23H/G68H (RSL-B6) in complex with cucurbit[7]uril and zinc | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SODIUM ION, ... | | Authors: | Guagnini, F, Engilberge, S, Flood, R.J, Ramberg, K.O, Crowley, P.B. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Metal-Mediated Protein-Cucurbituril Crystalline Architectures
Cryst.Growth Des., 2020
|
|
4MBO
 
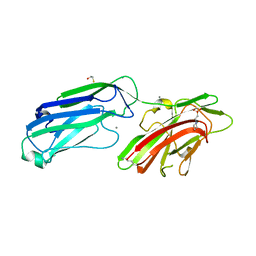 | | 1.65 Angstrom Crystal Structure of Serine-rich Repeat Adhesion Glycoprotein (Srr1) from Streptococcus agalactiae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
4MC1
 
 | | HIV protease in complex with SA526P | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4S)-4-tert-butyl-7-fluoro-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
4M3D
 
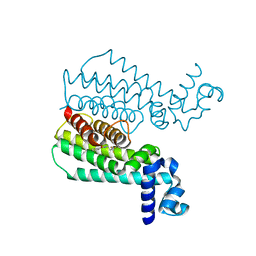 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-{3-[(phenylsulfonyl)amino]prop-1-yn-1-yl}-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
6M8M
 
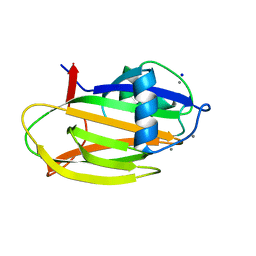 | | PA14 sugar-binding domain from RTX adhesin | | Descriptor: | CALCIUM ION, Putative large adhesion protein (Lap) involved in biofilm formation, SODIUM ION, ... | | Authors: | Vance, T.D.R, Conroy, B, Davies, P.L. | | Deposit date: | 2018-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and functional analysis of a bacterial adhesin sugar-binding domain.
Plos One, 14, 2019
|
|
4M3Q
 
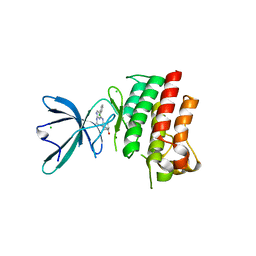 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1917 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Zhang, W, Zhang, D, Stashko, M.A, DeRyckere, D, Hunter, D, Kireev, D.B, Miley, M, Cummings, C, Lee, M, Norris-Drouin, J, Stewart, W.M, Sather, S, Zhou, Y, Kirkpatrick, G, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Pseudo-Cyclization through Intramolecular Hydrogen Bond Enables Discovery of Pyridine Substituted Pyrimidines as New Mer Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
6I6H
 
 | |
4MEQ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 5-methyl-triazolopyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-7-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4QT5
 
 | | Crystal Structure of 3BD10: A Monoclonal Antibody against the TSH Receptor | | Descriptor: | 3BD10 mouse monoclonal antibody, heavy chain, light chain | | Authors: | Hubbard, P.A, Chen, C.R, McLachlan, S.M, Rapoport, B, Murali, R. | | Deposit date: | 2014-07-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a TSH receptor monoclonal antibody: insight into Graves' disease pathogenesis.
Mol.Endocrinol., 29, 2015
|
|
6UYA
 
 | | Crystal structure of Compound 19 bound to IRAK4 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{2-[(2R)-2-fluoro-3-hydroxy-3-methylbutyl]-6-(morpholin-4-yl)-1-oxo-2,3-dihydro-1H-isoindol-5-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Kiefer, J.R, Bryan, M.C, Lupardus, P.J, Zarrin, A.A, Rajapaksa, N.S, Gobbi, A, Drobnick, J, Kolesnikov, A, Liang, J, Do, S. | | Deposit date: | 2019-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Potent Benzolactam IRAK4 Inhibitors with Robust in Vivo Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
4QU8
 
 | | Caspase-3 M61A V266H | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CHLORIDE ION, ... | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
3J82
 
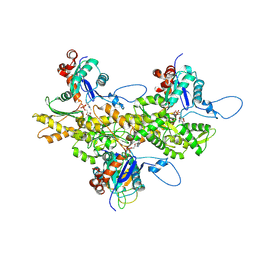 | | Electron cryo-microscopy of DNGR-1 in complex with F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Hanc, P, Fujii, T, Yamada, Y, Huotari, J, Schulz, O, Ahrens, S, Kjaer, S, Way, M, Namba, K, Reis e Sousa, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens.
Immunity, 42, 2015
|
|
4MHF
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Glucuronate, space group P21 | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit, alpha-D-glucopyranuronic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1LZV
 
 | | Site-Specific Mutant (Tyr7 replaced with His) of Human Carbonic Anhydrase II | | Descriptor: | Carbonic Anhydrase II, ZINC ION | | Authors: | Tu, C.K, Qian, M, An, H, Wadhwa, N.R, Duda, D.M, Yoshioka, C, Pathak, Y, McKenna, R, Laipis, P.J, Silverman, D.N. | | Deposit date: | 2002-06-11 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic analysis of multiple proton shuttles in the active site of human carbonic anhydrase.
J.Biol.Chem., 277, 2002
|
|
6MEK
 
 | |
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
1MAZ
 
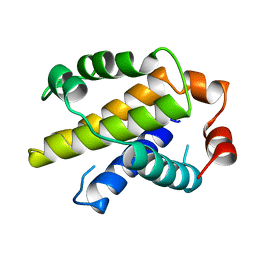 | | X-RAY STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH | | Descriptor: | Bcl-2-like protein 1 | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
6M8T
 
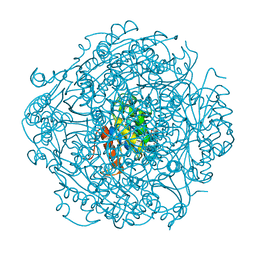 | | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, PHOSPHATE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutidnova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex
To Be Published
|
|
7AA2
 
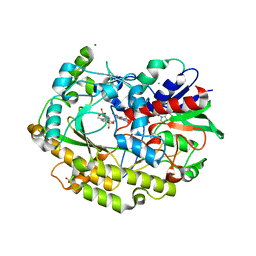 | | Chaetomium thermophilum FAD-dependent oxidoreductase in complex with ABTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6CMS
 
 | | Closed structure of active SHP2 mutant E76K bound to SHP099 inhibitor | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Padua, R.A.P, Sun, Y, Marko, I, Pitsawong, W, Kern, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun, 9, 2018
|
|
3KLN
 
 | | Vibrio cholerae VpsT | | Descriptor: | Transcriptional regulator, LuxR family | | Authors: | Krasteva, P.V, Navarro, V.A.S, Sondermann, H. | | Deposit date: | 2009-11-08 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Vibrio cholerae VpsT Regulates Matrix Production and Motility by Directly Sensing Cyclic di-GMP.
Science, 327, 2010
|
|
