6SCS
 
 | |
4RA9
 
 | | Crystal Structure of Conjoint Pyrococcus Furiosus L-asparaginase with Citrate | | Descriptor: | CITRATE ANION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Sharma, P, Tomar, R, Singh, S, Yadav, S.P.S, Ashish, Kundu, B. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and functional insights into an archaeal L-asparaginase obtained through the linker-less assembly of constituent domains.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7YWL
 
 | | Six DNA Helix Bundle nanopore - State 3 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
5MJ6
 
 | | Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Mpakali, A, Stratikos, E, Saridakis, E, Giastas, P. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Ligand-Induced Conformational Change of Insulin-Regulated Aminopeptidase: Insights on Catalytic Mechanism and Active Site Plasticity.
J. Med. Chem., 60, 2017
|
|
7YWI
 
 | | Six DNA duplex bundle nanopore - State 2 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7YWO
 
 | | Six DNA Helix Bundle nanopore - State 5 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
3KDP
 
 | | Crystal structure of the sodium-potassium pump | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, Na+/K+ ATPase gamma subunit transcript variant a, ... | | Authors: | Morth, J.P, Pedersen, B.P, Nissen, P. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the sodium-potassium pump.
Nature, 450, 2007
|
|
7YWN
 
 | | Six DNA Helix Bundle nanopore - State 4 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Orlova, E.V, Coveney, P, Howorka, S. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
4QUH
 
 | | Caspase-3 T140G | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
6PLA
 
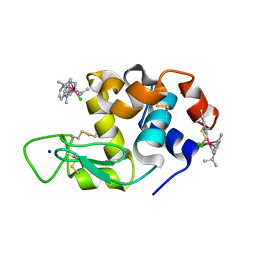 | | Adducts formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidazol-2-ylidene)(eta6-p-cymene)osmium(II) with HEWL | | Descriptor: | Lysozyme, OSMIUM ION, SODIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
7C1J
 
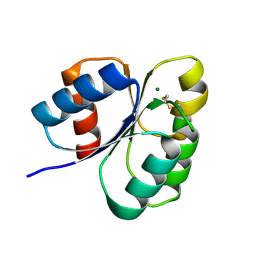 | | Crystal structure of the receiver domain of sensor histidine kinase PA1611 (PA1611REC) from Pseudomonas aeruginosa PAO1 with magnesium ion coordinated in the active site cleft | | Descriptor: | Histidine kinase, MAGNESIUM ION | | Authors: | Chen, S.K, Guan, H.H, Wu, P.H, Lin, L.T, Wu, M.C, Chang, H.Y, Chen, N.C, Lin, C.C, Chuankhayan, P, Huang, Y.C, Lin, P.J, Chen, C.J. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the histidine-containing phospho-transfer protein and receiver domain of sensor histidine kinase suggest a complex model in the two-component regulatory system in Pseudomonas aeruginosa
Iucrj, 7, 2020
|
|
4QWW
 
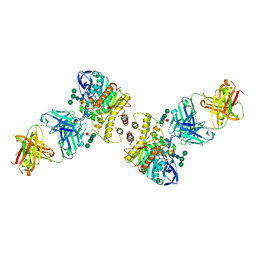 | | Crystal structure of the Fab410-BfAChE complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Renault, L, Marchot, P. | | Deposit date: | 2014-07-17 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Snake Venom Acetylcholinesterase in Complex with Inhibitory Antibody Fragment Fab410 Bound at the Peripheral Site: EVIDENCE FOR OPEN AND CLOSED STATES OF A BACK DOOR CHANNEL.
J.Biol.Chem., 290, 2015
|
|
3C0D
 
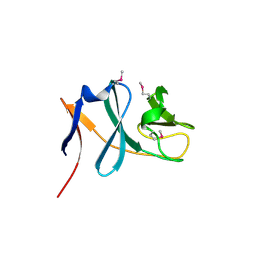 | | Crystal structure of the putative nitrite reductase NADPH (small subunit) oxidoreductase protein Q87HB1. Northeast Structural Genomics Consortium target VpR162 | | Descriptor: | Putative nitrite reductase NADPH (Small subunit) oxidoreductase protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Wang, D, Fang, Y, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-19 | | Release date: | 2008-03-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the putative nitrite reductase NADPH (small subunit) oxidoreductase protein Q87HB1.
To be Published
|
|
6YNF
 
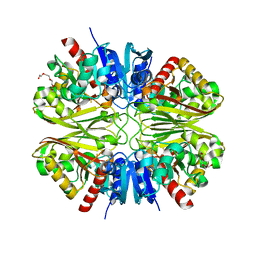 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 3 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
6SP2
 
 | | CryoEM structure of SERINC from Drosophila melanogaster | | Descriptor: | CARDIOLIPIN, Lauryl Maltose Neopentyl Glycol, Membrane protein TMS1d, ... | | Authors: | Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2019-08-30 | | Release date: | 2020-01-01 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | A bipartite structural organization defines the SERINC family of HIV-1 restriction factors.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8TXG
 
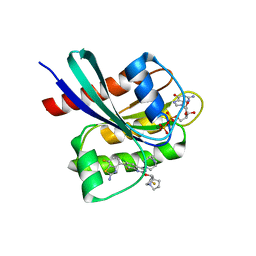 | | Crystal structure of KRAS G12D in complex with GDP and compound 8 | | Descriptor: | (4M)-4-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-7-fluoro-1,3-benzothiazol-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8C7B
 
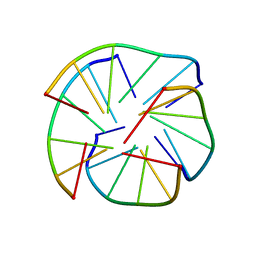 | |
4R5H
 
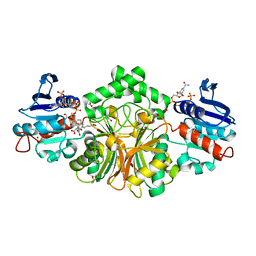 | | Crystal structure of sp-Aspartate-Semialdehyde-Dehydrogenase with Nicotinamide-Adenine-Dinucleotide-Phosphate and 3-carboxy-propenyl-phthalic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[(1E)-3-carboxyprop-1-en-1-yl]benzene-1,2-dicarboxylic acid, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Thangavelu, B, Bhansali, P, Viola, R.E. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5FJZ
 
 | |
6MP7
 
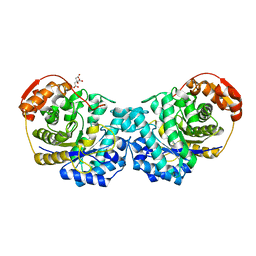 | | Crystal structure of the E257A mutant of BlMan5B in complex with GlcNAc (soaking) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BlMan5B, CITRATE ANION, ... | | Authors: | Lorizolla-Cordeiro, R, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2018-10-05 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | N-glycan Utilization by Bifidobacterium Gut Symbionts Involves a Specialist beta-Mannosidase.
J. Mol. Biol., 431, 2019
|
|
7PEM
 
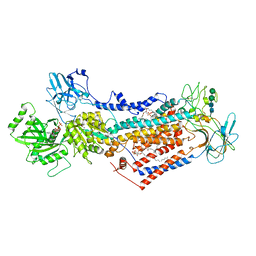 | | Cryo-EM structure of phophorylated Drs2p-Cdc50p in a PS and ATP-bound E2P state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Timcenko, M, Wang, Y, Lyons, J.A, Nissen, P, Lindorff-Larsen, K. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Substrate Transport and Specificity in a Phospholipid Flippase
To Be Published
|
|
8C7A
 
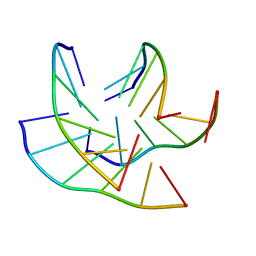 | |
4QQK
 
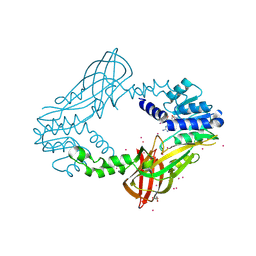 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) with GMS | | Descriptor: | (5S)-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}-N~6~-carbamimidoyl-L-lysine, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
5BX9
 
 | | Structure of PslG from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Baker, P, Little, D.J, Howell, P.L. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa Glycoside Hydrolase PslG Reveals That Its Levels Are Critical for Psl Polysaccharide Biosynthesis and Biofilm Formation.
J.Biol.Chem., 290, 2015
|
|
6MR2
 
 | |
