4OSK
 
 | | Crystal structure of TAL effector reveals the recognition between asparagine and guanine | | Descriptor: | DNA (5'-D(*AP*GP*TP*CP*TP*AP*GP*TP*AP*GP*TP*TP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*AP*AP*CP*TP*AP*CP*TP*AP*GP*AP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
6DLW
 
 | | Complement component polyC9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement component C9, beta-D-mannopyranose | | Authors: | Dunstone, M.A, Spicer, B.A, Law, R.H.P. | | Deposit date: | 2018-06-03 | | Release date: | 2018-09-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The first transmembrane region of complement component-9 acts as a brake on its self-assembly.
Nat Commun, 9, 2018
|
|
4OT0
 
 | | Crystal structure of the S505T mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
7Q9A
 
 | | MHC Class I A02 Allele presenting LLLGIGILVL, in complex with Mel5 TCR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Human Mel5 T Cell Receptor, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
6Z47
 
 | | Smooth muscle myosin shutdown state heads region | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin heavy chain 11, ... | | Authors: | Scarff, C.A, Carrington, G, Casas Mao, D, Chalovich, J.M, Knight, P.J, Ranson, N.A, Peckham, M. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structure of the shutdown state of myosin-2.
Nature, 588, 2020
|
|
4O6K
 
 | | The crystal structure of zebrafish IL-22 | | Descriptor: | Interleukin 22 | | Authors: | Siupka, P, Hamming, O.J, Fretaud, M, Luftalla, G, Levraud, J.P, Hartmann, R. | | Deposit date: | 2013-12-21 | | Release date: | 2014-09-10 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of zebrafish IL-22 reveals an evolutionary, conserved structure highly similar to that of human IL-22.
Genes Immun., 15, 2014
|
|
4OV0
 
 | | Structure of Bacteriorhdopsin Transferred from Amphipol A8-35 to a Lipidic Mesophase | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Polovinkin, V, Gushchin, I, Sintsov, M, Round, E, Balandin, T, Chervakov, P, Schevchenko, V, Utrobin, P, Popov, A, Borshchevskiy, V, Mishin, A, Kuklin, A, Willbold, D, Popot, J.L, Gordeliy, V. | | Deposit date: | 2014-02-19 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a membrane protein transferred from amphipol to a lipidic mesophase.
J.Membr.Biol., 247, 2014
|
|
5CVB
 
 | | Crystal structure of the type IX collagen NC2 hetero-trimerization domain with a guest fragment a1a1a1 of type I collagen | | Descriptor: | Collagen alpha-1(I) chain,Collagen alpha-1(IX) chain, Collagen alpha-1(I) chain,Collagen alpha-2(IX) chain, Collagen alpha-1(I) chain,Collagen alpha-3(IX) chain, ... | | Authors: | Boudko, S.P, Bachinger, H.P. | | Deposit date: | 2015-07-25 | | Release date: | 2016-08-10 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural insight for chain selection and stagger control in collagen.
Sci Rep, 6, 2016
|
|
7YAT
 
 | | CryoEM tetra protofilament structure of the hamster prion 108-144 fibril | | Descriptor: | Major prion protein | | Authors: | Chen, E.H.-L, Kao, S.-W, Lee, C.-H, Huang, J.Y.C, Chen, R.P.-Y, Wu, K.-P. | | Deposit date: | 2022-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 angstrom Cryo-EM Tetra-Protofilament Structure of the Hamster Prion 108-144 Fibril Reveals an Ordered Water Channel in the Center.
J.Am.Chem.Soc., 144, 2022
|
|
3UQN
 
 | | Crystal structure of dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Oxamic acid at 1.9 Angstrom resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, OXAMIC ACID | | Authors: | Singh, A, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Oxamic acid at 1.9 Angstrom resolution
To be Published
|
|
5K71
 
 | | apo Dbr1 | | Descriptor: | RNA lariat debranching enzyme, putative, SULFATE ION | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The RNA lariat debranching enzyme Dbr1: metal dependence and branched RNA co-crystal structures
Proc.Natl.Acad.Sci.USA, 2016
|
|
6DI4
 
 | |
7QT0
 
 | | Antibody FenAb136 - fentanyl complex | | Descriptor: | 2-[2-[2-[2-[[5-oxidanylidene-5-[2-[4-[phenyl(propanoyl)amino]piperidin-1-yl]ethylamino]pentanoyl]amino]ethanoylamino]ethanoylamino]ethanoylamino]ethanoic acid, Antibody heavy chain, Antibody light chain | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2022-01-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A trypanosome-derived immunotherapeutics platform elicits potent high-affinity antibodies, negating the effects of the synthetic opioid fentanyl.
Cell Rep, 42, 2023
|
|
5K5S
 
 | | Crystal structure of the active form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
5K73
 
 | | as-isolated Dbr1 with Fe(II) and Zn(II) | | Descriptor: | FE (II) ION, HYDROXIDE ION, RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The RNA lariat debranching enzyme Dbr1: metal dependence and branched RNA co-crystal structures
Proc.Natl.Acad.Sci.USA, 2016
|
|
6Z52
 
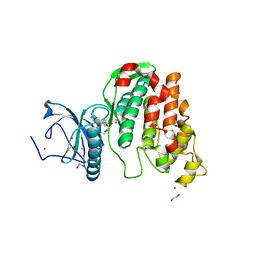 | | Crystal structure of CLK3 in complex with macrocycle ODS2003136 | | Descriptor: | 1,2-ETHANEDIOL, 11,15-dimethyl-6-(4-methylpiperazin-1-yl)-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2003136
To Be Published
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
3UH1
 
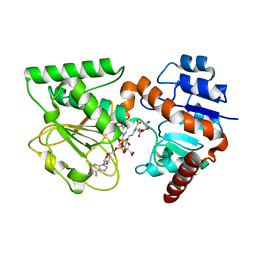 | | Crystal Structure of Saccharopine Dehydrogenase from Saccharomyces cerevisiae with bound saccharopine and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID, ... | | Authors: | Kumar, V.P, Thomas, L.M, Bobyk, K.D, Andi, B, West, A.H, Cook, P.F. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
6ZGG
 
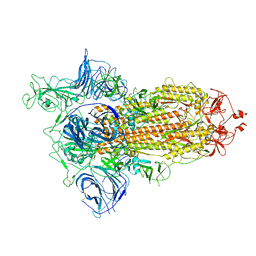 | |
5D9I
 
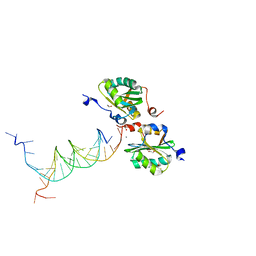 | | SV40 Large T antigen origin binding domain bound to artificial DNA fork | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Meinke, G, Bohm, A, Bullock, P.A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based analysis of the interaction between the simian virus 40 T-antigen origin binding domain and single-stranded DNA.
J. Virol., 85, 2011
|
|
5KH6
 
 | | SETDB1 in complex with a fragment candidate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Walker, J.R, Harding, R.J, Mader, P, Dobrovetsky, E, Dong, A, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Brown, P.J, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SETDB1 in complex with a fragment candidate
To be published
|
|
4OLJ
 
 | | Crystal structure of Arg119Gln mutant of Peptidyl-tRNA Hydrolase from Acinetobacter Baumannii at 1.49 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidyl-tRNA hydrolase | | Authors: | Sikarwar, J, Dube, D, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of Arg119Gln mutant of Peptidyl-tRNA hydrolase from Acinetobacter Baumannii at 1.49 A resolution
to be published
|
|
1W8J
 
 | |
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
4W1P
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 5.54 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-14 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
