3RUU
 
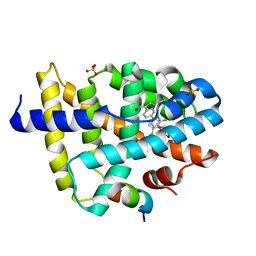 | | FXR with SRC1 and GSK237 | | Descriptor: | 6-(4-{[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazol-4-yl]methoxy}phenyl)-1H-indole-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Alternative replacements of the stilbene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4WV1
 
 | |
1B9E
 
 | | HUMAN INSULIN MUTANT SERB9GLU | | Descriptor: | PROTEIN (INSULIN) | | Authors: | Wang, D.C, Zeng, Z.H, Yao, Z.P, Li, H.M. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an insulin dimer in an orthorhombic crystal: the structure analysis of a human insulin mutant (B9 Ser-->Glu).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5KMW
 
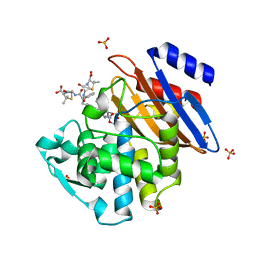 | | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex | | Descriptor: | Beta-lactamase Toho-1, OPEN FORM - PENICILLIN G, PENICILLIN G, ... | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Ginell, S.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex
to be published
|
|
5KPO
 
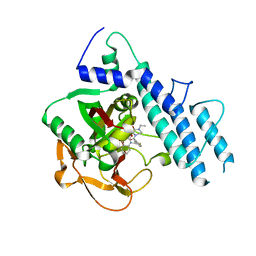 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[3-(4-ethyl-3-oxidanylidene-piperazin-1-yl)carbonyl-4-fluoranyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
1AX3
 
 | | SOLUTION NMR STRUCTURE OF B. SUBTILIS IIAGLC, 16 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE IIA DOMAIN | | Authors: | Chen, Y, Case, D.A, Reizer, J, Saier Junior, M.H, Wright, P.E. | | Deposit date: | 1997-10-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of Bacillus subtilis IIAglc.
Proteins, 31, 1998
|
|
6YN0
 
 | | Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity | | Descriptor: | Cell division protein FtsN, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Kerff, F, Terrak, M, Boes, A, Herman, H, Charlier, P. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The bacterial cell division protein fragment E FtsN binds to and activates the major peptidoglycan synthase PBP1b.
J.Biol.Chem., 295, 2020
|
|
3NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
1AW9
 
 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE III IN APO FORM | | Descriptor: | CADMIUM ION, GLUTATHIONE S-TRANSFERASE III | | Authors: | Neuefeind, T, Huber, R, Reinemer, P, Knaeblein, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, sequencing, crystallization and X-ray structure of glutathione S-transferase-III from Zea mays var. mutin: a leading enzyme in detoxification of maize herbicides.
J.Mol.Biol., 274, 1997
|
|
1WTV
 
 | | Hyperthermophile chromosomal protein SAC7D single mutant M29A in complex with DNA GTAATTAC | | Descriptor: | 5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
4D2P
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 7-({4-[(3-hydroxy-5-methoxyphenyl)amino]benzoyl}amino)-1,2,3,4-tetrahydroisoquinolinium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4WF4
 
 | | Crystal structure of E.Coli DsbA co-crystallised in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, Thiol:disulfide interchange protein | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5KUI
 
 | | Human mitochondrial calcium uniporter (residues 72-189) crystal structure with calcium. | | Descriptor: | Calcium uniporter protein, mitochondrial | | Authors: | Mok, M.C.Y, Lee, S.K, Junop, M.S, Stathopulos, P.B. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations.
Cell Chem Biol, 23, 2016
|
|
2HT0
 
 | | IHF bound to doubly nicked DNA | | Descriptor: | 5'-D(*CP*GP*GP*TP*GP*CP*AP*AP*CP*AP*AP*AP*T)-3', 5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3', 5'-D(*GP*GP*CP*CP*AP*AP*AP*AP*AP*AP*GP*CP*AP*TP*T)-3', ... | | Authors: | Swinger, K.K, Rice, P.A. | | Deposit date: | 2006-07-24 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based Analysis of HU-DNA Binding.
J.Mol.Biol., 365, 2007
|
|
1P4T
 
 | | Crystal structure of Neisserial surface protein A (NspA) | | Descriptor: | ETHANOLAMINE, PENTAETHYLENE GLYCOL MONODECYL ETHER, SULFATE ION, ... | | Authors: | Vandeputte-Rutten, L, Bos, M.P, Tommassen, J, Gros, P. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Neisserial Surface Protein A (NspA),
a conserved outer membrane protein with vaccine potential
J.Biol.Chem., 278, 2003
|
|
8IIC
 
 | |
1P5N
 
 | |
3O2X
 
 | | MMP-13 in complex with selective tetrazole core inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Collagenase 3, ... | | Authors: | Kiefer, J.R, Barta, T.E, Becker, D.P, Mathis, K.J, Williams, J. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MMP-13 in complex with selective tetrazole core inhibitor
To be Published
|
|
4D9O
 
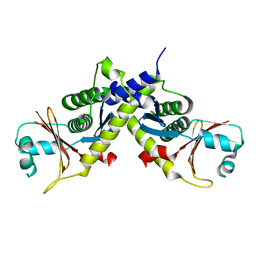 | | Structure of ebolavirus protein VP24 from Reston | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Zhang, A.P.P. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold
Plos Pathog., 8, 2012
|
|
2RIC
 
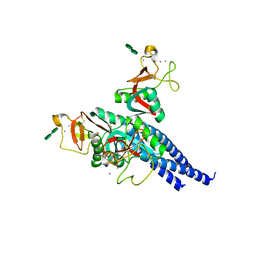 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with L-glycero-D-manno-heptopyranosyl-(1-3)-L-glycero-D-manno-heptopyranose | | Descriptor: | CALCIUM ION, L-glycero-alpha-D-manno-heptopyranose, L-glycero-alpha-D-manno-heptopyranose-(1-3)-L-glycero-alpha-D-manno-heptopyranose, ... | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
1AV3
 
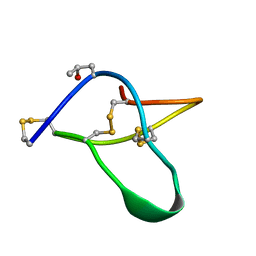 | | POTASSIUM CHANNEL BLOCKER KAPPA CONOTOXIN PVIIA FROM C. PURPURASCENS, NMR, 20 STRUCTURES | | Descriptor: | Kappa-conotoxin PVIIA | | Authors: | Scanlon, M.J, Naranjo, D, Thomas, L, Alewood, P.F, Lewis, R.J, Craik, D.J. | | Deposit date: | 1997-09-24 | | Release date: | 1998-10-14 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and proposed binding mechanism of a novel potassium channel toxin kappa-conotoxin PVIIA.
Structure, 5, 1997
|
|
2HNY
 
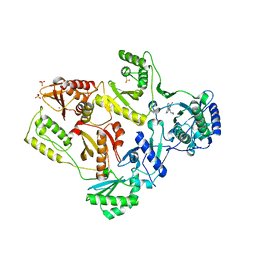 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
351D
 
 | |
2MDA
 
 | |
3E6S
 
 | |
