8F65
 
 | |
6YM4
 
 | | Crystal structure of BAY-297 with PIP4K2A | | Descriptor: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YKG
 
 | |
4QY4
 
 | | Crystal structure of the bromodomain of human SMARCA2 | | Descriptor: | SMARCA2 protein, ZINC ION | | Authors: | Tallant, C, Savitsky, P, Nunez-Alonso, G, Fonseca, M, Krojer, T, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of the bromodomain of human SMARCA2
To be Published
|
|
6NKA
 
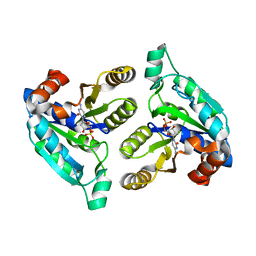 | |
3CVP
 
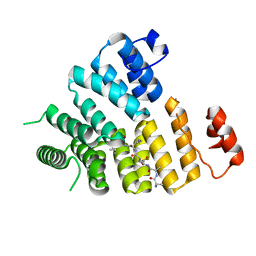 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (10-SKL) | | Descriptor: | 10-SKL PTS1 peptide Ac-GTLSNRASKL, Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
8EFZ
 
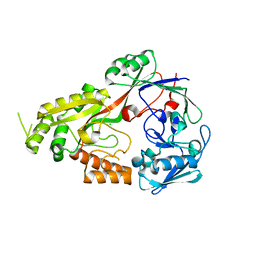 | | Crystal structure of CcNikZ-II, apoprotein | | Descriptor: | CHLORIDE ION, Extracellular solute-binding protein family 5 | | Authors: | Stogios, P.J, Evdokimova, E, Diep, P, Yakunin, A, Mahadevan, K, Savchenko, A. | | Deposit date: | 2022-09-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of CcNikZ-II, apoprotein
To Be Published
|
|
5KCJ
 
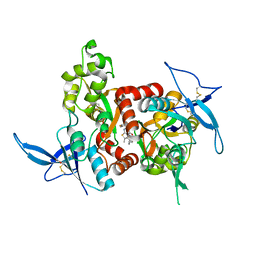 | | Structure of the human GluN1/GluN2A LBD in complex with GNE6901 | | Descriptor: | 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
8EJU
 
 | | The crystal structure of Pseudomonas putida PcaR | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Transcription regulatory protein (Pca regulon), ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR
To Be Published
|
|
6GRR
 
 | | Crystal structure of Escherichia coli amine oxidase mutant I342F/E573Q | | Descriptor: | Amine oxidase, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Gaule, T.G, Smith, M.A, Tych, K.M, Pirrat, P, Trinh, C.H, Pearson, A.R, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2018-06-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxygen Activation Switch in the Copper Amine Oxidase of Escherichia coli.
Biochemistry, 57, 2018
|
|
5YI2
 
 | | Structure of Lactococcus lactis ZitR, wild type in complex with DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*A)-3'), ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
6Y1T
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a Trp51 to S-Trp51 modification | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
6Y2Y
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with Trp51 to S-Trp51 and Trp191Phe modifications | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
8A12
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to Mg.ATP-gamma-S | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2022-05-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
5NIF
 
 | | Yeast 20S proteasome in complex with Blm-pep activator | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Witkowska, J, Grudnik, P, Golik, P, Dubin, G, Jankowska, E. | | Deposit date: | 2017-03-23 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a low molecular weight activator Blm-pep with yeast 20S proteasome - insights into the enzyme activation mechanism.
Sci Rep, 7, 2017
|
|
6WXO
 
 | | De novo TIM barrel-ferredoxin fold fusion homodimer with 2-histidine 2-glutamate centre TFD-HE | | Descriptor: | GLYCEROL, SULFATE ION, TFD-HE | | Authors: | Caldwell, S.J, Zeymer, C, Haydon, I.C, Huang, P, Hilvert, D, Baker, D. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5CDR
 
 | | 2.65 structure of S.aureus DNA gyrase and artificially nicked DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3'), DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
8FBW
 
 | | Crystal structure of SIV-1 V2 antibody NCI05 in complex with a V2 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, T, Kwong, P.D, Van Wazer, D.J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Effect of Passive Administration of Monoclonal Antibodies Recognizing Simian Immunodeficiency Virus (SIV) V2 in CH59-Like Coil/Helical or beta-Sheet Conformations on Time of SIV mac251 Acquisition.
J.Virol., 97, 2023
|
|
8FMA
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C11 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FNW
 
 | | Structure of RdrA-RdrB complex from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, Archaeal ATPase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
6NTI
 
 | | Neutron/X-ray crystal structure of AAC-VIa bound to kanamycin b | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6Z1Z
 
 | | Structure of the anti-CD9 nanobody 4C8 | | Descriptor: | Nanobody 4C8 | | Authors: | Neviani, N, Oosterheert, W, Pearce, N.M, Lutz, M, Kroon-Batenburg, L.M.J, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
5KE2
 
 | | Crystal structure of SETDB1 Tudor domain in complex with inhibitor XST06472A | | Descriptor: | (3~{S})-~{N}-~{tert}-butyl-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with inhibitor xst06472a
to be published
|
|
8J0A
 
 | | Robust design of effective allosteric activator UbV R4 for Rsp5 E3 ligase using the machine-learning tool ProteinMPNN | | Descriptor: | SULFATE ION, Ubiquitin variant R4 | | Authors: | Lin, Y.-F, Hsieh, Y.-J, Kao, H.-W, Ko, T.-P, Wu, K.-P. | | Deposit date: | 2023-04-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Robust Design of Effective Allosteric Activators for Rsp5 E3 Ligase Using the Machine Learning Tool ProteinMPNN.
Acs Synth Biol, 12, 2023
|
|
