1V3D
 
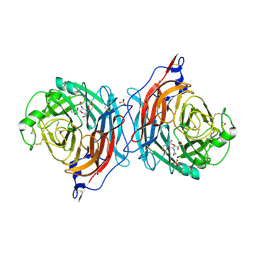 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with NEU5AC2EN | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1YG4
 
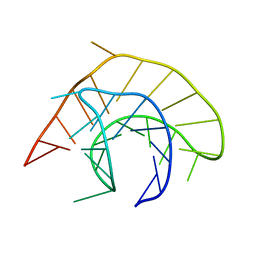 | |
4DP6
 
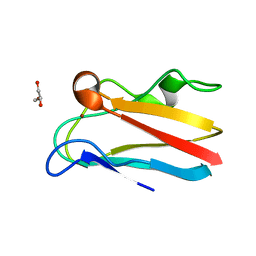 | | The 1.67 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 8.0 | | Descriptor: | COPPER (I) ION, GLYCEROL, Plastocyanin B, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
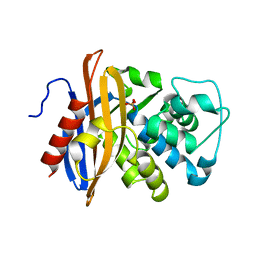
1MBL
 
 | |
6IMG
 
 | | Solution Structure of Bicyclic Peptide pb-13 | | Descriptor: | (ACE)-GLY-CYS-PRO-CYS-ILE-TRP-PRO-GLU-LEU-CYS-PRO-TRP-ILE-ARG-SER-CYS-(NH2) | | Authors: | Yao, H, Lin, P, Zha, J, Zha, M, Zhao, Y, Wu, C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Method: | SOLUTION NMR | | Cite: | Ordered and Isomerically Stable Bicyclic Peptide Scaffolds Constrained through Cystine Bridges and Proline Turns.
Chembiochem, 20, 2019
|
|
1CDT
 
 | | CARDIOTOXIN V4/II FROM NAJA MOSSAMBICA MOSSAMBICA: THE REFINED CRYSTAL STRUCTURE | | Descriptor: | CARDIOTOXIN VII4, PHOSPHATE ION | | Authors: | Rees, B, Bilwes, A, Samama, J.P, Moras, D. | | Deposit date: | 1990-05-17 | | Release date: | 1991-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cardiotoxin VII4 from Naja mossambica mossambica. The refined crystal structure.
J.Mol.Biol., 214, 1990
|
|
1Y82
 
 | | Conserved hypothetical protein Pfu-367848-001 from Pyrococcus furiosus | | Descriptor: | UNKNOWN ATOM OR ION, hypothetical protein | | Authors: | Horanyi, P, Tempel, W, Habel, J, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-367848-001 from Pyrococcus furiosus
To be published
|
|
4AW8
 
 | | X-ray structure of ZinT from Salmonella enterica in complex with zinc ion and PEG | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Metal-binding protein ZinT, SODIUM ION, ... | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
1KZO
 
 | | PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH FARNESYLATED K-RAS4B PEPTIDE PRODUCT AND FARNESYL DIPHOSPHATE SUBSTRATE BOUND SIMULTANEOUSLY | | Descriptor: | ACETIC ACID, FARNESYL, FARNESYL DIPHOSPHATE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2002-02-07 | | Release date: | 2002-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Reaction Path of Protein Farnesyltransferase at Atomic Resolution
Nature, 419, 2002
|
|
2X4W
 
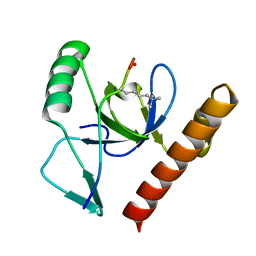 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | FORMIC ACID, HISTONE H3.2, PEREGRIN | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1C5F
 
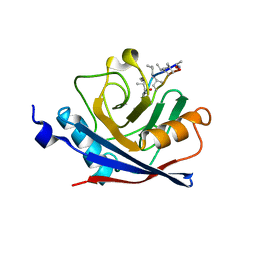 | | CRYSTAL STRUCTURE OF THE CYCLOPHILIN-LIKE DOMAIN FROM BRUGIA MALAYI COMPLEXED WITH CYCLOSPORIN A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE 1 | | Authors: | Ellis, P.J, Carlow, C.K.S, Ma, D, Kuhn, P. | | Deposit date: | 1999-11-22 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of the Complex of Brugia Malayi Cyclophilin and Cyclosporin A.
Biochemistry, 39, 2000
|
|
6JH8
 
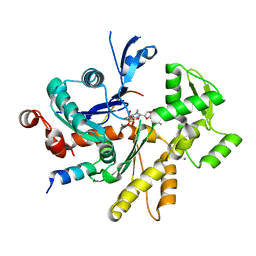 | |
6JHH
 
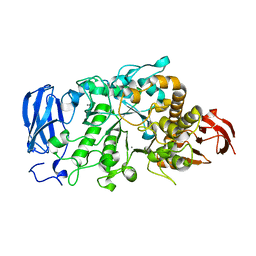 | | Crystal structure of mutant D350A of Pullulanase from Paenibacillus barengoltzii complexed with maltotriose | | Descriptor: | CALCIUM ION, Pulullanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | Crystal structure of mutant D350A of Pullulanase from Paenibacillus barengoltzii complexed with maltotriose
To Be Published
|
|
1Y8V
 
 | | Crystal structure of the A-DNA GCGTAT*CGC with a 2'-O-propyl Thymidine (T*) | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(P2T)P*AP*CP*GP*C)-3' | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Influence of Stereoelectronic Effects on the Biophysical Properties of Oligonucleotides: Comprehensive Analysis of the RNA Affinity, Nuclease Resistance, and Crystal Structure of Ten 2'-O-Ribonucleic Acid Modifications.
Biochemistry, 44, 2005
|
|
3GHE
 
 | |
3F6U
 
 | | Crystal structure of human Activated Protein C (APC) complexed with PPACK | | Descriptor: | CALCIUM ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Schmidt, A.E, Padmanabhan, K, Underwood, M.C, Bode, W, Mather, T, Bajaj, S.P. | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermodynamic linkage between the S1 site, the Na+ site, and the Ca2+ site in the protease domain of human activated protein C (APC).
J.Biol.Chem., 277, 2002
|
|
3GEY
 
 | | Crystal structure of human poly(ADP-ribose) polymerase 15, catalytic fragment in complex with an inhibitor Pj34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Poly [ADP-ribose] polymerase 15 | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
5XEU
 
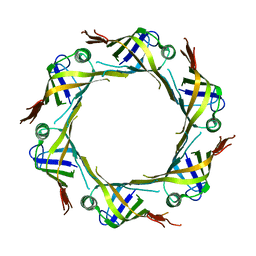 | | crystal structure of Hcp2 from Salmonella typhimurium | | Descriptor: | Hcp1 family type VI secretion system effector | | Authors: | Lin, Q.P, Gao, Z.Q, Zhang, H. | | Deposit date: | 2017-04-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the putative cytoplasmic protein STM0279 (Hcp2) from Salmonella typhimurium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5RH6
 
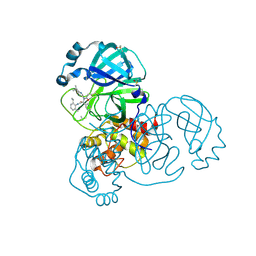 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z4439011588 (Mpro-x2703) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]-N-[6-(propan-2-yl)pyridin-3-yl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
4B1L
 
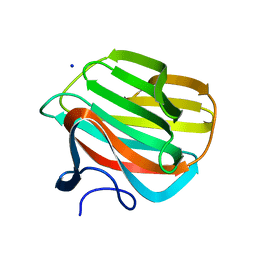 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, beta-D-fructofuranose | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WRU
 
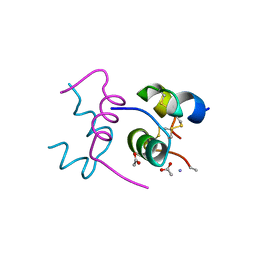 | | Semi-synthetic highly active analogue of human insulin NMeAlaB26-DTI- NH2 | | Descriptor: | ACETATE ION, INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1YB9
 
 | | Crystal structure of the A-DNA GCGTAT*CGC with a 2'-O-[2-(N,N-dimethylaminooxy)ethyl] Thymidine (T*) | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2OT)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharan, M. | | Deposit date: | 2004-12-20 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Probing the Influence of Stereoelectronic Effects on the Biophysical Properties of Oligonucleotides: Comprehensive Analysis of the RNA Affinity, Nuclease Resistance, and Crystal Structure of Ten 2'-O-Ribonucleic Acid Modifications.
Biochemistry, 44, 2005
|
|
3G5L
 
 | | Crystal structure of putative S-adenosylmethionine dependent methyltransferase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Putative S-adenosylmethionine dependent methyltransferase | | Authors: | Patskovsky, Y, Sampathkumar, P, Gilmore, M, Miller, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of S-Adenosylmethionine Dependent Methyltransferase from Listeria Monocytogenes
To be Published
|
|
3F9K
 
 | | Two domain fragment of HIV-2 integrase in complex with LEDGF IBD | | Descriptor: | Integrase, MAGNESIUM ION, PC4 and SFRS1-interacting protein, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75
Plos Pathog., 5, 2009
|
|
4O5X
 
 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine. | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
