5SB8
 
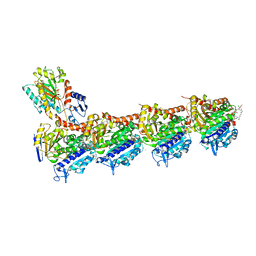 | | Tubulin-maytansinoid-3-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-6-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaene-8,23-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
3ZQR
 
 | | NMePheB25 insulin analogue crystal structure | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
5LEB
 
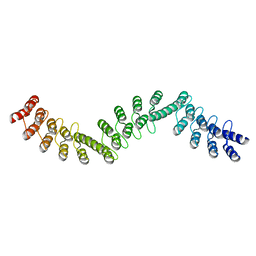 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_06_D12_06_D12 | | Descriptor: | DDD_D12_06_D12_06_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
1XRN
 
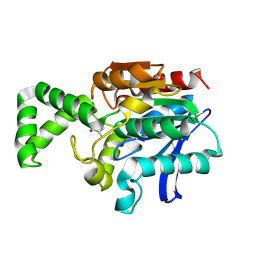 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Phe-Ala | | Descriptor: | ALANINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
5SBD
 
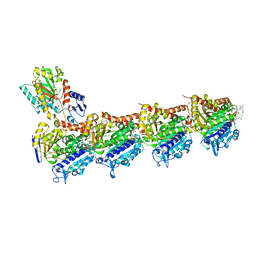 | | Tubulin-maytansinoid-5b-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21(25)-hexaen-6-yl acetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
3ZHU
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, second post-decarboxylation intermediate from 2-oxoadipate | | Descriptor: | (5R)-5-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-5-(2-{[(phosphonatooxy)phosphinato]oxy}ethyl)-1,3-thiazol-3-ium-2-yl}-5-hydroxypentanoate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
1BSX
 
 | | STRUCTURE AND SPECIFICITY OF NUCLEAR RECEPTOR-COACTIVATOR INTERACTIONS | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, PROTEIN (GRIP1), PROTEIN (THYROID HORMONE RECEPTOR BETA) | | Authors: | Wagner, R.L, Darimont, B.D, Apriletti, J.W, Stallcup, M.R, Kushner, P.J, Baxter, J.D, Fletterick, R.J, Yamamoto, K.R. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and specificity of nuclear receptor-coactivator interactions.
Genes Dev., 12, 1998
|
|
5SBA
 
 | | Tubulin-maytansinoid-4b-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl acetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
7PG7
 
 | |
5SBE
 
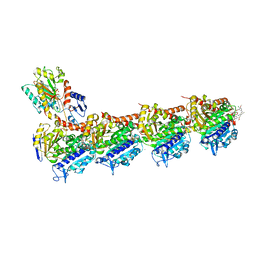 | | Tubulin-maytansinoid-5c-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaen-6-yl hept-6-ynoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
2W80
 
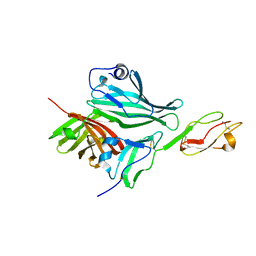 | | Structure of a complex between Neisseria meningitidis factor H binding protein and CCPs 6-7 of human complement factor H | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Schneider, M.C, Prosser, B.E, Caesar, J.J.E, Kugelberg, E, Li, S, Zhang, Q, Quoraishi, S, Lovett, J.E, Deane, J.E, Sim, R.B, Roversi, P, Johnson, S, Tang, C.M, Lea, S.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Nature, 458, 2009
|
|
6Q6Y
 
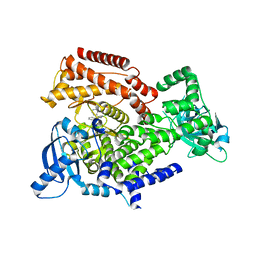 | | PI3K delta in complex with N(2chloro5phenylpyridin3yl)benzenesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-(2-chloranyl-5-phenyl-pyridin-3-yl)benzenesulfonamide | | Authors: | Convery, M.A, Rowland, P, Down, K, Barton, N. | | Deposit date: | 2018-12-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Potent, Efficient, and Selective Inhibitors of Phosphoinositide 3-Kinase delta through a Deconstruction and Regrowth Approach.
J.Med.Chem., 61, 2018
|
|
5SBB
 
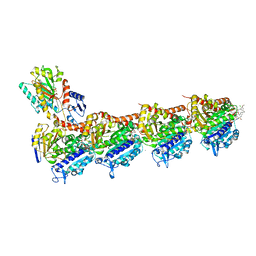 | | Tubulin-maytansinoid-4c-complex | | Descriptor: | (1R,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl heptanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBC
 
 | | Tubulin-maytansinoid-5a-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaen-6-yl phenylacetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5X4W
 
 | | Mutant human thymidylate synthase A191K crystallized in a sulfate-free condition | | Descriptor: | PHOSPHATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5SB9
 
 | | Tubulin-maytansinoid-4a-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl phenylacetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
4XSK
 
 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
7TR3
 
 | | CaKip3[2-482] - AMP-PNP in complex with a dolastatin-10-stabilized tubulin ring | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Hunter, B, Allingham, J.S, Sosa, H. | | Deposit date: | 2022-01-27 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Kinesin-8-specific loop-2 controls the dual activities of the motor domain according to tubulin protofilament shape.
Nat Commun, 13, 2022
|
|
7PKO
 
 | | CryoEM structure of Rotavirus NSP2 | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1C01
 
 | | SOLUTION STRUCTURE OF MIAMP1, A PLANT ANTIMICROBIAL PROTEIN | | Descriptor: | ANTIMICROBIAL PEPTIDE 1 | | Authors: | McManus, A.M, Nielsen, K.J, Marcus, J.P, Harrison, S.J, Green, J.L, Manners, J.M, Craik, D.J. | | Deposit date: | 1999-07-13 | | Release date: | 2000-07-19 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | MiAMP1, a novel protein from Macadamia integrifolia adopts a Greek key beta-barrel fold unique amongst plant antimicrobial proteins.
J.Mol.Biol., 293, 1999
|
|
4DYH
 
 | | Crystal structure of glycosylated Lipase from Humicola lanuginosa at 2 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Singh, A, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-02-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glycosylated Lipase from Humicola lanuginosa at 2 Angstrom resolution
TO BE PUBLISHED
|
|
2NXB
 
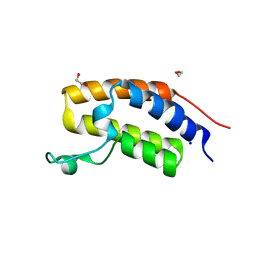 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, SODIUM ION | | Authors: | Filippakopoulos, P, Bullock, A, Cooper, C, Keates, K, Berridge, G, Pike, A, Bunkoczi, G, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
1JCS
 
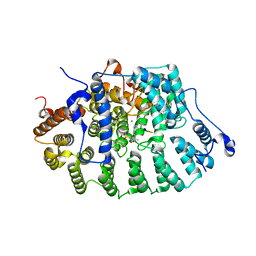 | | CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH THE PEPTIDE SUBSTRATE TKCVFM AND AN ANALOG OF FARNESYL DIPHOSPHATE | | Descriptor: | ACETIC ACID, PROTEIN FARNESYLTRANSFERASE, ALPHA SUBUNIT, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4XUV
 
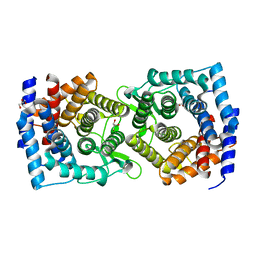 | | Crystal structure of a glycoside hydrolase family 105 (GH105) enzyme from Thielavia terrestris | | Descriptor: | GLYCEROL, Glycoside hydrolase family 105 protein | | Authors: | Stogios, P.J, Xu, X, Cui, H, Yim, V, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0496 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 105 (GH105) enzyme from Thielavia terrestris
To Be Published
|
|
3MM9
 
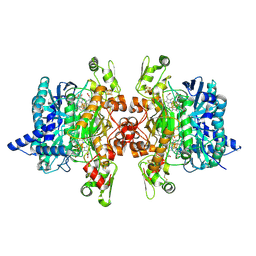 | | Dissimilatory sulfite reductase nitrite complex | | Descriptor: | IRON/SULFUR CLUSTER, NITRITE ION, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
