4UCD
 
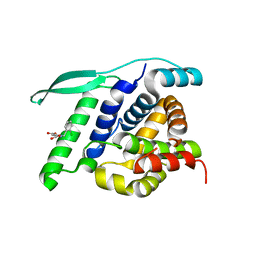 | | N-terminal globular domain of the RSV Nucleoprotein in complex with the Nucleoprotein Phosphoprotein interaction inhibitor M81 | | Descriptor: | 1-[(2-chlorophenyl)methyl]pyrazole-3,5-dicarboxylic acid, NUCLEOPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UCA
 
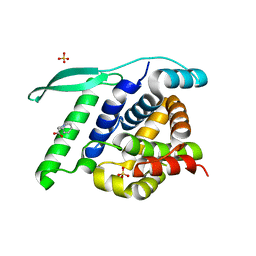 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal peptide of the Phosphoprotein | | Descriptor: | NUCLEOPROTEIN, PHOSPHOSPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UEW
 
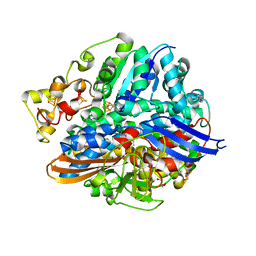 | | Structure of H2-treated anaerobically purified D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | [NiFe]-hydrogenases revisited: nickel-carboxamido bond formation in a variant with accrued O2-tolerance and a tentative re-interpretation of Ni-SI states.
Metallomics, 7, 2015
|
|
4UPE
 
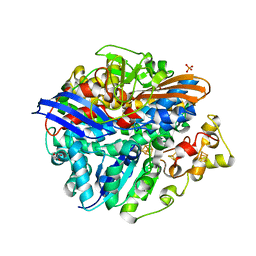 | | Structure of the unready Ni-A state of the S499C mutant of D. fructosovorans NiFe-hydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4V48
 
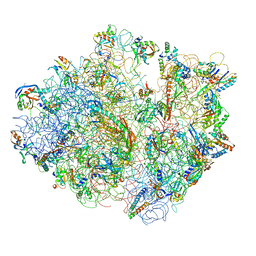 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
4ZLU
 
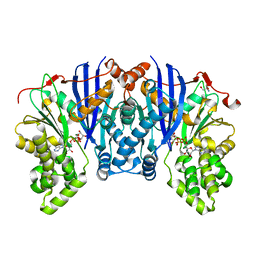 | |
4N3M
 
 | | Joint neutron/X-ray structure of urate oxidase in complex with 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-02-05 | | Last modified: | 2018-06-13 | | Method: | NEUTRON DIFFRACTION (1.919 Å), X-RAY DIFFRACTION | | Cite: | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
6TM8
 
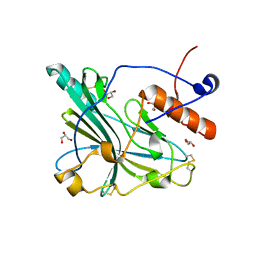 | | Crystal structure of glycoprotein D of Equine Herpesvirus Type 4 | | Descriptor: | Envelope glycoprotein D, GLYCEROL | | Authors: | Kremling, V, Loll, B, Osterrieder, N, Wahl, M, Dahmani, I, Chiantia, P, Azab, W. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycoprotein D of equine alphaherpesviruses reveal potential binding sites to the entry receptor MHC-I.
Front Microbiol, 14, 2023
|
|
4Z8K
 
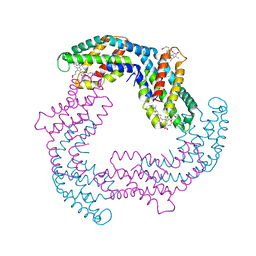 | |
3F6H
 
 | | Crystal structure of the regulatory domain of LiCMS in complexed with isoleucine - type III | | Descriptor: | Alpha-isopropylmalate synthase, ISOLEUCINE, ZINC ION | | Authors: | Zhang, P, Ma, J, Zhao, G, Ding, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans
Biochem.J., 421, 2009
|
|
6TPV
 
 | | Crystal structures of FNIII domain one and two of the human leucocyte common antigen-related protein, LAR | | Descriptor: | IMIDAZOLE, Receptor-type tyrosine-protein phosphatase F | | Authors: | Vilstrup, J.P, Thirup, S.S, Simonsen, A, Birkefeldt, T, Strandbygaard, D. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structures of fragments of the human leucocyte common antigen-related protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4ZKR
 
 | |
4ZL5
 
 | |
3F92
 
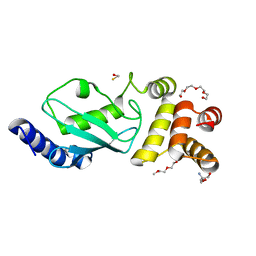 | | Crystal structure of ubiquitin-conjugating enzyme E2-25kDa (Huntington Interacting Protein 2) M172A mutant crystallized at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Wilson, R.C, Hughes, R.C, Flatt, J.W, Meehan, E.J, Ng, J.D, Twigg, P.D. | | Deposit date: | 2008-11-13 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4ZOG
 
 | |
3FN3
 
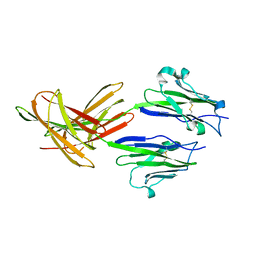 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
4Z1P
 
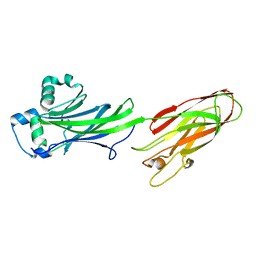 | | BspA_C_mut | | Descriptor: | Cell wall surface anchor protein | | Authors: | Race, P, Rego, S. | | Deposit date: | 2015-03-27 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BspA_C_mut
To Be Published
|
|
9BER
 
 | | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Cell Rep, 43, 2024
|
|
4RVE
 
 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*TP*AP*TP*CP*CP*C)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilso, K.S. | | Deposit date: | 1993-02-18 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
4Z38
 
 | |
3FK6
 
 | | Crystal structure of TetR triple mutant (H64K, S135L, S138I) | | Descriptor: | Tetracycline repressor protein class B from transposon Tn10, Tetracycline repressor protein class D | | Authors: | Klieber, M.A, Scholz, O, Lochner, S, Gmeiner, P, Hillen, W, Muller, Y.A. | | Deposit date: | 2008-12-16 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origins for selectivity and specificity in an engineered bacterial repressor-inducer pair.
Febs J., 276, 2009
|
|
4ZCJ
 
 | |
4ZDG
 
 | |
4ZFI
 
 | | Structure of Mdm2 with low molecular weight inhibitor | | Descriptor: | (5S)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxy-1-methyl-1,5-dihydro-2H-pyrrol-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Zak, K.M, Twarda-Clapa, A, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-21 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
6TD4
 
 | | IRF4 DNA-binding domain surface entropy mutant apo structure | | Descriptor: | CHLORIDE ION, Interferon regulatory factor 4 | | Authors: | Tucker, J.A, Martin, M.P, Wang, L.Z, Jennings, C, Heath, R. | | Deposit date: | 2019-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Cancer-associated mutations in the IRF4 DNA-binding domain confer no disadvantage in DNA-binding affinity and may increase transcriptional activity
To Be Published
|
|
