5AEI
 
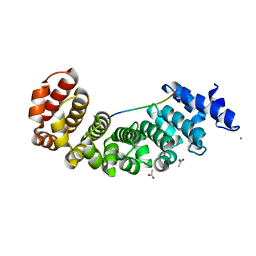 | | Designed Armadillo repeat protein YIIIM5AII in complex with peptide (KR)5 | | Descriptor: | ACETATE ION, CALCIUM ION, DESIGNED ARMADILLO REPEAT PROTEIN YIIIM5AII, ... | | Authors: | Hansen, S, Tremmel, D, Madhurantakam, C, Reichen, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2015-08-31 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Energetic Contributions of a Designed Modular Peptide-Binding Protein with Picomolar Affinity.
J.Am.Chem.Soc., 138, 2016
|
|
5OPB
 
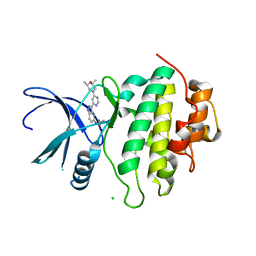 | | Structure of CHK1 10-pt. mutant complex with indazole LRRK2 inhibitor | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
8EK6
 
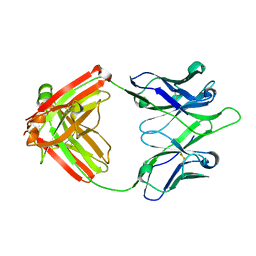 | | UCA Y35N (unbound) Fab from CH65-CH67 lineage | | Descriptor: | UCA Fab heavy chain, UCA Fab light chain | | Authors: | Maurer, D.P, Schmidt, A.G. | | Deposit date: | 2022-09-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Hierarchical sequence-affinity landscapes shape the evolution of breadth in an anti-influenza receptor binding site antibody.
Elife, 12, 2023
|
|
2R9Y
 
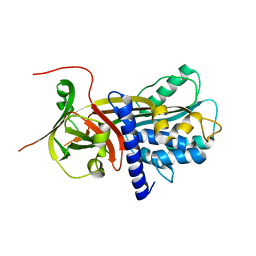 | | Structure of antiplasmin | | Descriptor: | Alpha-2-antiplasmin | | Authors: | Law, R.H.P, Sofian, T, Kan, W.T, Horvath, A.J, Hitchen, C.R, Langendorf, C.G, Buckle, A.M, Whisstock, J.C, Coughlin, P.B. | | Deposit date: | 2007-09-14 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray crystal structure of the fibrinolysis inhibitor {alpha}2-antiplasmin
Blood, 111, 2008
|
|
6DU6
 
 | | Crystal structure of the pyruvate kinase (PK1) from the mosquito Aedes aegypti | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Pyruvate kinase | | Authors: | Pizarro, J.C, Scaraffia, P.Y, Petchampai, N, Murillo-Solano, C. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.513 Å) | | Cite: | Distinctive regulatory properties of pyruvate kinase 1 from Aedes aegypti mosquitoes.
Insect Biochem. Mol. Biol., 104, 2018
|
|
5OQ8
 
 | | Structure of CHK1 12-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
8EKH
 
 | | I-2 Y35N H35N (unbound) Fab from CH65-CH67 lineage | | Descriptor: | I-2 Fab heavy chain, I-2 Fab light chain | | Authors: | Maurer, D.P, Schmidt, A.G. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hierarchical sequence-affinity landscapes shape the evolution of breadth in an anti-influenza receptor binding site antibody.
Elife, 12, 2023
|
|
1M25
 
 | | STRUCTURE OF SYNTHETIC 26-MER PEPTIDE CONTAINING 145-169 SHEEP PRION PROTEIN SEGMENT AND C-TERMINAL CYSTEINE IN TFE SOLUTION | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Megy, S, Bertho, G, Kozin, S.A, Coadou, G, Debey, P, Hoa, G.H, Girault, J.-P. | | Deposit date: | 2002-06-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Possible role of region 152-156 in the structural duality of a peptide fragment from sheep prion protein
Protein Sci., 13, 2004
|
|
2RBO
 
 | |
4O6Z
 
 | | Crystal structure of serine hydroxymethyltransferase with covalently bound PLP Schiff-base from Plasmodium falciparum | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2013-12-24 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The structure of Plasmodium falciparum serine hydroxymethyltransferase reveals a novel redox switch that regulates its activities.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8EQH
 
 | |
8BQ7
 
 | |
1SF2
 
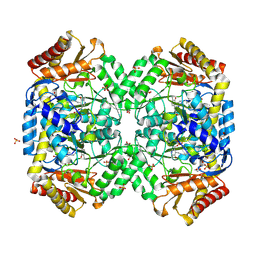 | | Structure of E. coli gamma-aminobutyrate aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
2IU3
 
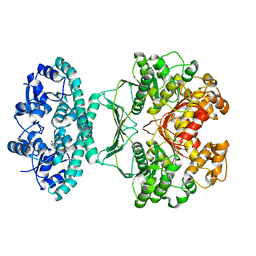 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase.
J. Biol. Chem., 282, 2007
|
|
3UKI
 
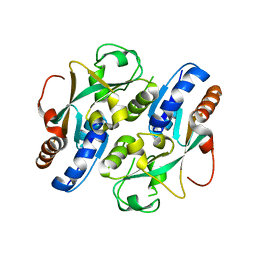 | | Crystal structure of reduced OxyR from Porphyromonas gingivalis | | Descriptor: | OxyR | | Authors: | Svintradze, D.V, Wright, H.T, Collazo-Santiago, E.A, Lewis, J.P. | | Deposit date: | 2011-11-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structures of the Porphyromonas gingivalis OxyR regulatory domain explain differences in expression of the OxyR regulon in Escherichia coli and P. gingivalis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4C6Z
 
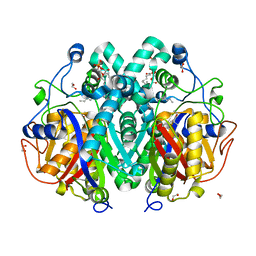 | | Crystal structure of M. tuberculosis C171Q KasA in complex with TLM3 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(10R)-10-methyloctadecanoyl]oxy}propyl phosphate, (5R)-3-ethyl-4-hydroxy-5-methyl-5-[(1E)-2-methylbuta-1,3-dien-1-yl]thiophen-2(5H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Schiebel, J, Kapilashrami, K, Fekete, A, Bommineni, G.R, Schaefer, C.M, Mueller, M.J, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-09-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the Recognition of Mycolic Acid Precursors by Kasa, a Condensing Enzyme and Drug Target from Mycobacterium Tuberculosis
J.Biol.Chem., 288, 2013
|
|
2IU0
 
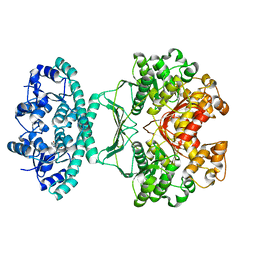 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
8BP6
 
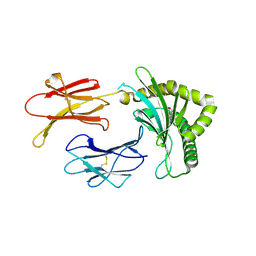 | | Structure of MHC-class I related molecule MR1 with bound M3Ade | | Descriptor: | (1R,5S)-8-(9H-purin-6-yl)-2-oxa-8-azabicyclo[3.3.1]nona-3,6-diene-4,6-dicarbaldehyde, Beta-2-microglobulin,Major histocompatibility complex class I-related gene protein | | Authors: | Berloffa, G, Jakob, R.P, Maier, T. | | Deposit date: | 2022-11-16 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The carbonyl nucleoside adduct M3Ade stabilizes MR1 and activates MR1-restricted self- and tumor-reactive T cells
To Be Published
|
|
2RIF
 
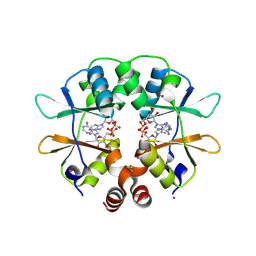 | | CBS domain protein PAE2072 from Pyrobaculum aerophilum complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CESIUM ION, Conserved protein with 2 CBS domains | | Authors: | Lee, T.M, King, N.P, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures and Functional Implications of an AMP-Binding Cystathionine beta-Synthase Domain Protein from a Hyperthermophilic Archaeon.
J.Mol.Biol., 380, 2008
|
|
4K2Z
 
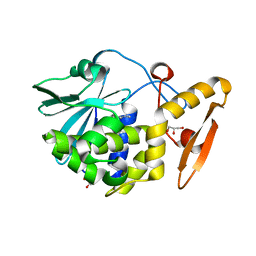 | | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with Methylethylamine at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, METHYLETHYLAMINE, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with Methylethylamine at 1.80 A resolution
To be Published
|
|
8EX1
 
 | |
3QRJ
 
 | | The crystal structure of human abl1 kinase domain T315I mutant in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
2J67
 
 | | The TIR domain of human Toll-Like Receptor 10 (TLR10) | | Descriptor: | TOLL LIKE RECEPTOR 10 | | Authors: | Stenmark, P, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Magnusdottir, A, Nilsson, M.E, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2006-09-26 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Human Toll-Like Receptor 10 Cytoplasmic Domain Reveals a Putative Signaling Dimer.
J.Biol.Chem., 283, 2008
|
|
3MLZ
 
 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with a VI191 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5OYH
 
 | | crystal structure of the catalytic core of a rhodopsin-guanylyl cyclase with converted specificity in complex with ATPalphaS | | Descriptor: | ADENOSINE-5'-SP-ALPHA-THIO-TRIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Broser, M, Scheib, U, Hegemann, P. | | Deposit date: | 2017-09-09 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Rhodopsin-cyclases for photocontrol of cGMP/cAMP and 2.3 angstrom structure of the adenylyl cyclase domain.
Nat Commun, 9, 2018
|
|
