4EAM
 
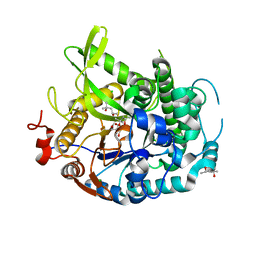 | | 1.70A resolution structure of apo beta-glycosidase (W33G) from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, ... | | Authors: | Lovell, S, Battaile, K.P, Deckert, K, Brunner, L.C, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designing allosteric control into enzymes by chemical rescue of structure.
J.Am.Chem.Soc., 134, 2012
|
|
8RNV
 
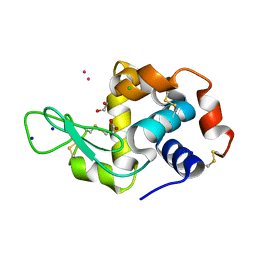 | | Hen Egg White Lysozyme soaked with cis-Ru(DMSO)4Cl2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
8RNX
 
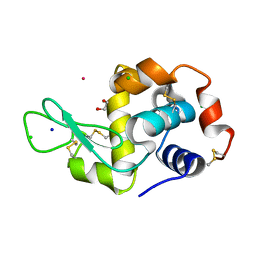 | | Hen Egg White Lysozyme soaked with [HIsq][trans-RuCl4(DMSO)(Isq)] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
6KZ8
 
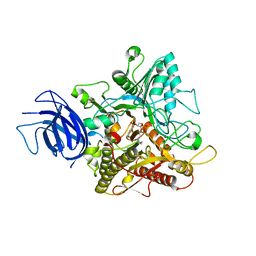 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
9CPO
 
 | |
2Q0N
 
 | | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Turnbull, A, Papagrigoriou, E, Pike, A.W, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-22 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide.
To be Published
|
|
4ECB
 
 | | Chimeric GST Containing Inserts of Kininogen Peptides | | Descriptor: | chimeric protein between GSHKT10 and domain 5 of kininogen-1 | | Authors: | Amber, A.B, Sergei, M.M, Yi, P, Rita, R, Xiaoping, Q, Marianne, P.-C, William, C.M, Vivien, Y, Keith, R.M, Anton, A.K. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chimeric glutathione S-transferases containing inserts of kininogen peptides: potential novel protein therapeutics.
J.Biol.Chem., 287, 2012
|
|
2PR4
 
 | | Crystal Structure of Fab' from the HIV-1 Neutralizing Antibody 2F5 | | Descriptor: | nmAb 2F5 Fab' Heavy Chain, nmAb 2F5 Fab' light Chain | | Authors: | Bryson, S, Julien, J.-P, Pai, E.F. | | Deposit date: | 2007-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
8RIJ
 
 | | Discovery of the first orally bioavailable ADAMTS7 inhibitor BAY-9835 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schafer, M, Meibom, D, Wasnaire, P, Beyer, K, Broehl, A, Cancho-Grande, Y, Elowe, N, Henninger, K, Johannes, S, Jungmann, N, Krainz, T, Lindner, N, Maassen, S, MacDonald, B, Menshykau, D, Mittendorf, J, Sanchez, G, Stefan, E, Torge, A, Xing, Y, Zubov, D. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | BAY-9835: Discovery of the First Orally Bioavailable ADAMTS7 Inhibitor.
J.Med.Chem., 67, 2024
|
|
4EGK
 
 | | Human Hsp90-alpha ATPase domain bound to Radicicol | | Descriptor: | Heat shock protein HSP 90-alpha, RADICICOL | | Authors: | Chen, W.J, Wood, S.P. | | Deposit date: | 2012-03-31 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment screening using capillary electrophoresis (CEfrag) for hit identification of heat shock protein 90 ATPase inhibitors.
J Biomol Screen, 17, 2012
|
|
4ERX
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with diethylene glycol at 2.5 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Singh, N, Sinha, M, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with diethylene glycol at 2.5 Angstrom resolution
To be Published
|
|
4ESW
 
 | | Crystal structure of C. albicans Thi5 H66G mutant | | Descriptor: | CITRIC ACID, Pyrimidine biosynthesis enzyme THI13 | | Authors: | Fenwick, M.K, Huang, S, Zhang, Y, Lai, R, Hazra, A, Rajashankar, K, Philmus, B, Kinsland, C, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Thiamin pyrimidine biosynthesis in Candida albicans : a remarkable reaction between histidine and pyridoxal phosphate.
J.Am.Chem.Soc., 134, 2012
|
|
4E4A
 
 | |
4ER6
 
 | | Crystal structure of human DOT1L in complex with inhibitor SGC0946 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, BROMIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
2PW2
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKWKSL | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, peptide epitope | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2007-05-10 | | Release date: | 2007-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications.
J.Virol., 83, 2009
|
|
6L77
 
 | | Crystal structure of MS5 from Brassica napus | | Descriptor: | MS5a | | Authors: | Wang, X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the meiosis-related protein MS5 reveals non-canonical papain enhancement by cystatin-like folds.
Febs Lett., 594, 2020
|
|
4E8E
 
 | | Structural characterization of Bombyx mori glutathione transferase BmGSTD1 | | Descriptor: | Glutathione S-transferase | | Authors: | Tan, X, Ma, X.X, Hu, X.M, Chen, Q.M, Zhao, P, Xia, Q.Y, Zhou, C.Z. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural characterization of Bombyx mori glutathione transferase BmGSTD1
To be Published
|
|
4EAD
 
 | | Thymidine phosphorylase from E.coli with 3'-azido-2'-fluoro-dideoxyuridine | | Descriptor: | 2',3'-dideoxy-2'-fluoro-3'-triaza-1,2-dien-2-ium-1-yluridine, GLYCEROL, SULFATE ION, ... | | Authors: | Timofeev, V.I, Abramchik, Y.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thymidine phosphorylase from E.coli with 3'-azido-2'-fluoro-dideoxyuridine
To be Published
|
|
4E75
 
 | | Structure of LpxD from Acinetobacter baumannii at 2.85A resolution (P21 form) | | Descriptor: | UDP-3-O-acylglucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure determination of LpxD from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4EA3
 
 | | Structure of the N/OFQ Opioid Receptor in Complex with a Peptide Mimetic | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-benzyl-N-[3-(1'H,3H-spiro[2-benzofuran-1,4'-piperidin]-1'-yl)propyl]-D-prolinamide, ... | | Authors: | Thompson, A.A, Liu, W, Chun, E, Katritch, V, Wu, H, Vardy, E, Huang, X.P, Trapella, C, Guerrini, R, Calo, G, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic.
Nature, 485, 2012
|
|
4EM2
 
 | |
2PMW
 
 | | The Crystal Structure of Proprotein convertase subtilisin kexin type 9 (PCSK9) | | Descriptor: | Proprotein convertase subtilisin/kexin type 9, SULFATE ION | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of PCSK9: A Regulator of Plasma LDL-Cholesterol.
Structure, 15, 2007
|
|
4EFE
 
 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
4EPR
 
 | | Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-Mediated Activation. | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, Q, Burke, J.P, Phan, J, Burns, M.C, Olejniczak, E.T, Waterson, A.G, Lee, T, Rossanese, O.W, Fesik, S.W. | | Deposit date: | 2012-04-17 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-Mediated Activation.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
