3NBB
 
 | | Crystal structure of mutant Y305F expressed in E. coli in the copper amine oxidase from hansenula polymorpha | | Descriptor: | COPPER (II) ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
5LBA
 
 | | Crystal structure of human RECQL5 helicase in complex with DSPL fragment(1-cyclohexyl-3-(oxolan-2-ylmethyl)urea, SGC - Diamond XChem I04-1 fragment screening. | | Descriptor: | 1-cyclohexyl-3-[[(2~{R})-oxolan-2-yl]methyl]urea, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, ... | | Authors: | Newman, J.A, Aitkenhead, H, Talon, R, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human RECQL5 helicase in complex with 3D fragment (1-cyclohexyl-3-(oxolan-2-ylmethyl)urea)
To be published
|
|
4ZFJ
 
 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC, apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Amidohydrolase EgtC | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
4QMC
 
 | | Crystal structure of complex formed between phospholipase A2 and Biotin-sulfoxide at 1.09 A Resolution | | Descriptor: | ACETATE ION, BIOTIN-D-SULFOXIDE, GLYCEROL, ... | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal structure of complex formed between phospholipase A2 and Biotin-sulfoxide at 1.09 A Resolution
To be published
|
|
3NJ8
 
 | |
2OIJ
 
 | | HIV-1 subtype B DIS RNA extended duplex AuCl3 soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', GOLD 3+ ION, POTASSIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2007-01-11 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes.
Nucleic Acids Res., 31, 2003
|
|
2KRV
 
 | |
5KSP
 
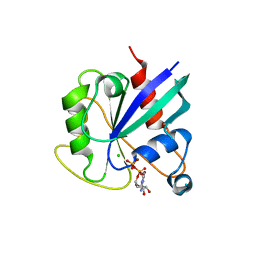 | | hMiro1 C-domain GDP Complex C2221 Crystal Form | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5GJA
 
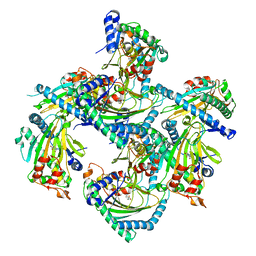 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with 2-PA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRIDINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
1BRJ
 
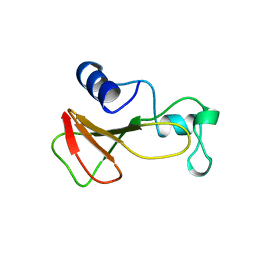 | | BARNASE MUTANT WITH ILE 88 REPLACED BY ALA | | Descriptor: | BARNASE, ZINC ION | | Authors: | Cramer, P.C, Buckle, A, Fersht, A. | | Deposit date: | 1995-03-09 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
1UZ6
 
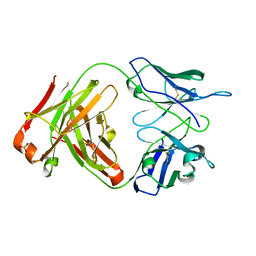 | | anti-Lewis X Fab fragment uncomplexed | | Descriptor: | IGG FAB (IGG3, KAPPA) HEAVY CHAIN 291-2G3-A, KAPPA) LIGHT CHAIN 291-2G3-A, ... | | Authors: | Van Roon, A.M.M, Pannu, N.S, De Vrind, J.P.M, Hokke, C.H, Deelder, A.M, Van Der marel, G.A, Van Boom, J.H, Abrahams, J.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an Anti-Lewis X Fab Fragment in Complex with its Lewis X Antigen
Structure, 12, 2004
|
|
5E4H
 
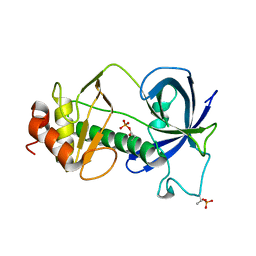 | |
3NFH
 
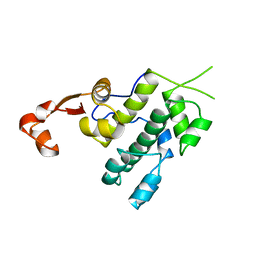 | | Crystal structure of tandem winged helix domain of RNA polymerase I subunit A49 (P4) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
6N0X
 
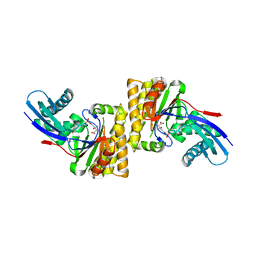 | |
6CP3
 
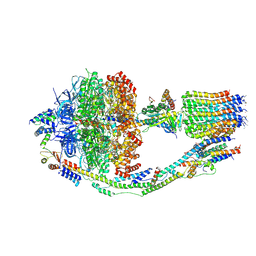 | | Monomer yeast ATP synthase (F1Fo) reconstituted in nanodisc with inhibitor of oligomycin bound. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
1BRH
 
 | | BARNASE MUTANT WITH LEU 14 REPLACED BY ALA | | Descriptor: | BARNASE, ZINC ION | | Authors: | Cramer, P.C, Buckle, A, Fersht, A. | | Deposit date: | 1995-03-09 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
1UVA
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | MYRISTIC ACID, NONSPECIFIC LIPID TRANSFER PROTEIN 1 | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
4CBY
 
 | | Design, synthesis, and biological evaluation of potent and selective Class IIa HDAC inhibitors as a potential therapy for Huntington's disease | | Descriptor: | (1R,2R,3R)-2-[4-(1,3-oxazol-5-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, SODIUM ION, ... | | Authors: | Burli, R.W, Luckhurst, C.A, Aziz, O, Matthews, K.L, Yates, D, Lyons, K.A, Beconi, M, McAllister, G, Breccia, P, Stott, A.J, Penrose, S.D, Wall, M, Lamers, M, Leonard, P, Mueller, I, Richardson, C.M, Jarvis, R, Stones, L, Hughes, S, Wishart, G, Haughan, A.F, O'Connell, C, Mead, T, McNeil, H, Vann, J, Mangette, J, Maillard, M, Beaumont, V, Munoz-Sanjuan, I, Dominguez, C. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease.
J. Med. Chem., 56, 2013
|
|
5E9J
 
 | |
6N4G
 
 | |
1IGN
 
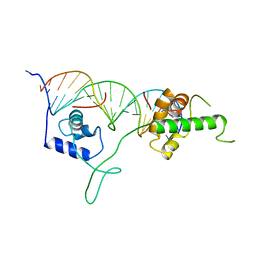 | | DNA-BINDING DOMAIN OF RAP1 IN COMPLEX WITH TELOMERIC DNA SITE | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*AP*CP*AP*CP*CP*CP*AP*CP*AP*CP*AP*CP*C P*AP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*GP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G P*CP*G)-3'), PROTEIN (RAP1) | | Authors: | Koenig, P, Giraldo, R, Chapman, L, Rhodes, D. | | Deposit date: | 1996-02-29 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA.
Cell(Cambridge,Mass.), 85, 1996
|
|
4NJK
 
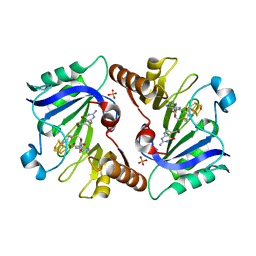 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 7-carboxy-7-deazaguanine, and Mg2+ | | Descriptor: | 2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
1DGK
 
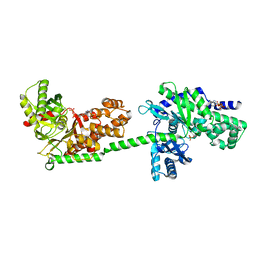 | | MUTANT MONOMER OF RECOMBINANT HUMAN HEXOKINASE TYPE I WITH GLUCOSE AND ADP IN THE ACTIVE SITE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE TYPE I, PHOSPHATE ION, ... | | Authors: | Aleshin, A.E, Liu, X, Kirby, C, Bourenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation.
J.Mol.Biol., 296, 2000
|
|
1UVB
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
2WZW
 
 | | Crystal structure of the FMN-dependent nitroreductase NfnB from Mycobacterium smegmatis in complex with NADPH | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NFNB PROTEIN, ... | | Authors: | Bellinzoni, M, Manina, G, Riccardi, G, Alzari, P.M. | | Deposit date: | 2009-12-03 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Characterization of the Mycobacterium Smegmatis Nitroreductase Nfnb, and its Role in Benzothiazinone Resistance
Mol.Microbiol., 77, 2010
|
|
