8HWZ
 
 | |
5TS5
 
 | |
5C7L
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase apo form | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
3ZK9
 
 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA D280N IN THE METAL-FREE, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
3P0G
 
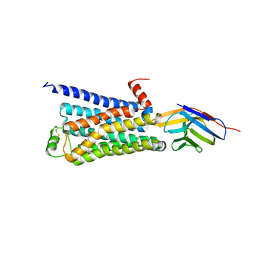 | | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Rasmussen, S.G.F, Choi, H.-J, Fung, J.J, Pardon, E, Casarosa, P, Chae, P.S, DeVree, B.T, Rosenbaum, D.M, Thian, F.S, Kobilka, T.S, Schnapp, A, Konetzki, I, Sunahara, R.K, Gellman, S.H, Pautsch, A, Steyaert, J, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Nature, 469, 2011
|
|
1AIK
 
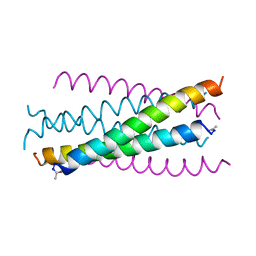 | | HIV GP41 CORE STRUCTURE | | Descriptor: | HIV-1 GP41 GLYCOPROTEIN | | Authors: | Chan, D.C, Fass, D, Berger, J.M, Kim, P.S. | | Deposit date: | 1997-04-20 | | Release date: | 1997-06-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Core structure of gp41 from the HIV envelope glycoprotein.
Cell(Cambridge,Mass.), 89, 1997
|
|
2FJS
 
 | |
5YPU
 
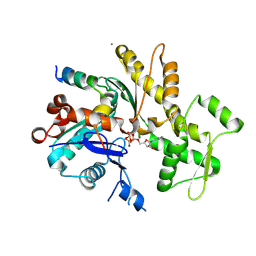 | | Crystal structure of an actin monomer in complex with the nucleator Cordon-Bleu MET72NLE WH2-motif peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Scipion, C.P.M, Wongsantichon, J, Ferrer, F.J, Yuen, T.Y, Robinson, R.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the roles of divalent cations in actin polymerization and activation of ATP hydrolysis
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4BR7
 
 | | Legionella pneumophila NTPDase1 crystal form I, open, AMPNP complex | | Descriptor: | 5'-O-[(R)-hydroxy(phosphonoamino)phosphoryl]adenosine, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4XUM
 
 | |
4BVM
 
 | | The peripheral membrane protein P2 from human myelin at atomic resolution | | Descriptor: | CITRIC ACID, MYELIN P2 PROTEIN, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Yadav, R.P, Kursula, P. | | Deposit date: | 2013-06-26 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Atomic Resolution View Into the Structure-Function Relationships of the Human Myelin Peripheral Membrane Protein P2
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5MOA
 
 | | ABA RECEPTOR FROM TOMATO, SlPYL1 | | Descriptor: | SlPYL1 | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5UGS
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT501 | | Descriptor: | 5-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]-2-(2-methylphenoxy)phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
3F5M
 
 | | Crystal Structure of ATP-Bound Phosphofructokinase from Trypanosoma brucei | | Descriptor: | 6-phospho-1-fructokinase (ATP-dependent phosphofructokinase), ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | McNae, I.W, Martinez-Oyanedel, J, Keillor, J.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of ATP-bound phosphofructokinase from Trypanosoma brucei reveals conformational transitions different from those of other phosphofructokinases.
J.Mol.Biol., 385, 2009
|
|
1A3X
 
 | | PYRUVATE KINASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH PG, MN2+ AND K+ | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Jurica, M.S, Mesecar, A, Heath, P.J, Shi, W, Nowak, T, Stoddard, B.L. | | Deposit date: | 1998-01-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate.
Structure, 6, 1998
|
|
2O5X
 
 | |
2JR4
 
 | | NMR Solution Structure of the Anticodon of E.coli TRNA-VAL3 With no Modifications | | Descriptor: | 5'-R(*CP*CP*UP*CP*CP*CP*UP*UP*AP*CP*AP*AP*GP*GP*AP*GP*G)-3' | | Authors: | Vendeix, F.A.P, Dziergowska, A, Gustilo, E.M, Graham, W.D, Sproat, B, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2007-06-20 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding.
Biochemistry, 47, 2008
|
|
1A6A
 
 | | THE STRUCTURE OF AN INTERMEDIATE IN CLASS II MHC MATURATION: CLIP BOUND TO HLA-DR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ghosh, P, Amaya, M, Mellins, E, Wiley, D.C. | | Deposit date: | 1998-02-22 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3.
Nature, 378, 1995
|
|
4BRO
 
 | |
1AH1
 
 | | CTLA-4, NMR, 20 STRUCTURES | | Descriptor: | CTLA-4, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Metzler, W.J, Bajorath, J, Fenderson, W, Shaw, S.-Y, Peach, R, Constantine, K.L, Naemura, J, Leytze, G, Lavoie, T.B, Mueller, L, Linsley, P.S. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human CTLA-4 and delineation of a CD80/CD86 binding site conserved in CD28.
Nat.Struct.Biol., 4, 1997
|
|
6Z3O
 
 | |
2JT3
 
 | | Solution Structure of F153W cardiac troponin C | | Descriptor: | Troponin C | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
1AKO
 
 | | EXONUCLEASE III FROM ESCHERICHIA COLI | | Descriptor: | EXONUCLEASE III | | Authors: | Mol, C.D, Kuo, C.-F, Thayer, M.M, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the multifunctional DNA-repair enzyme exonuclease III.
Nature, 374, 1995
|
|
1YCY
 
 | | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein | | Authors: | Huang, L, Liu, Z.-J, Lee, D, Tempel, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus
To be published
|
|
1KKP
 
 | | Crystal Structure of Serine Hydroxymethyltransferase complexed with Serine | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, Serine Hydroxymethyltransferase | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
