8CNJ
 
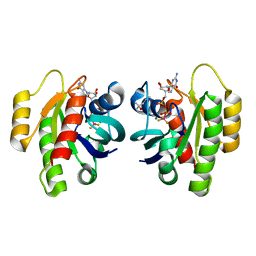 | | HRas(1-166) in complex with GDP and BeF3- | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, BERYLLIUM TRIFLUORIDE ION, GTPase HRas, ... | | 著者 | Baumann, P, Jin, Y. | | 登録日 | 2023-02-23 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Far-reaching effects of tyrosine64 phosphorylation on Ras revealed with BeF 3 - complexes.
Commun Chem, 7, 2024
|
|
2MCT
 
 | | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus | | 分子名称: | Uncharacterized protein | | 著者 | Martin, B.T, Serrano, P, Geralt, M, Dutta, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2013-08-27 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus.
To be Published
|
|
6NN9
 
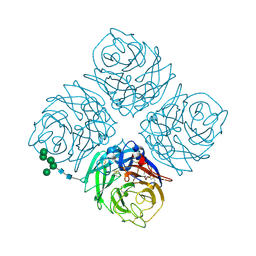 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | 著者 | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | 登録日 | 1991-03-28 | | 公開日 | 1992-07-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
6NNH
 
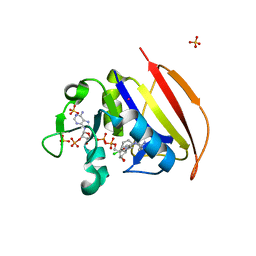 | | Structure of Closed state of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and cycloguanil | | 分子名称: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, COBALT (II) ION, Dihydrofolate reductase, ... | | 著者 | Giudice, J.H.P, Ribeiro, J.A, Dias, M.V.B. | | 登録日 | 2019-01-15 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.523 Å) | | 主引用文献 | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8C0V
 
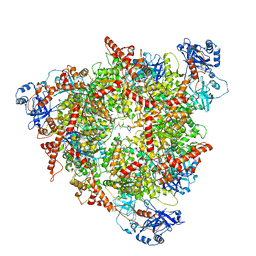 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in single seam state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | 登録日 | 2022-12-19 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
3QF6
 
 | | Neutron structure of type-III Antifreeze Protein allows the reconstruction of AFP-ice interface | | 分子名称: | Type-3 ice-structuring protein HPLC 12 | | 著者 | Howard, E.I, Blakeley, M.P, Haertlein, M, Petit-Haertlein, I, Mitschler, A, Fisher, S.J, Cousido-Siah, A, Salvay, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A. | | 登録日 | 2011-01-21 | | 公開日 | 2011-06-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å) | | 主引用文献 | Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface.
J.Mol.Recognit., 24, 2011
|
|
8C0W
 
 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | 登録日 | 2022-12-19 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
5KT8
 
 | |
1P7A
 
 | | Solution Structure of the Third Zinc Finger from BKLF | | 分子名称: | Kruppel-like factor 3, ZINC ION | | 著者 | Simpson, R.J.Y, Cram, E.D, Czolij, R, Matthews, J.M, Crossley, M, Mackay, J.P. | | 登録日 | 2003-04-30 | | 公開日 | 2003-12-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions.
J.Biol.Chem., 278, 2003
|
|
3QP2
 
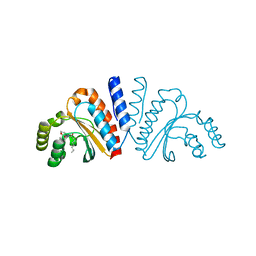 | | Crystal structure of CviR ligand-binding domain bound to C8-HSL | | 分子名称: | CviR transcriptional regulator, N-(2-OXOTETRAHYDROFURAN-3-YL)OCTANAMIDE | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.638 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
7O60
 
 | | Crystal structure of human myelin protein P2 at room temperature from joint X-ray and neutron refinement. | | 分子名称: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | 著者 | Laulumaa, S, Blakeley, M.P, Kursula, P. | | 登録日 | 2021-04-09 | | 公開日 | 2021-09-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
Febs J., 288, 2021
|
|
3VCA
 
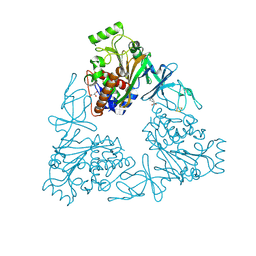 | | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman and UV-visible Spectroscopic Analysis of a Rieske-type Demethylase | | 分子名称: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | 著者 | Daughtry, K.D, Xiao, Y, Stoner-Ma, D, Cho, E, Orville, A.M, Liu, P, Allen, K.N. | | 登録日 | 2012-01-03 | | 公開日 | 2012-02-08 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman, and UV-Visible Spectroscopic Analysis of a Rieske-Type Demethylase.
J.Am.Chem.Soc., 134, 2012
|
|
7EZV
 
 | |
7EY4
 
 | |
3QBJ
 
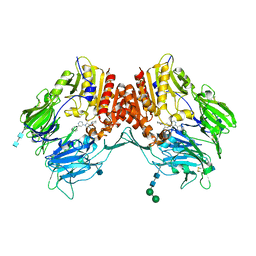 | | Crystal structure of dipeptidyl peptidase IV in complex with inhibitor | | 分子名称: | 1-[(3S,4S)-4-amino-1-(6-phenylpyrimidin-4-yl)pyrrolidin-3-yl]piperidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Liu, S.P. | | 登録日 | 2011-01-13 | | 公開日 | 2012-01-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Crystal structure of dipeptidyl peptidase IV in complex with inhibitor
To be Published
|
|
3VST
 
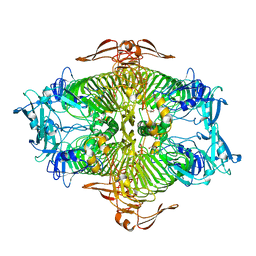 | | The complex structure of XylC with Tris | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Xylosidase | | 著者 | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | 登録日 | 2012-05-09 | | 公開日 | 2013-02-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
2M26
 
 | |
3QDZ
 
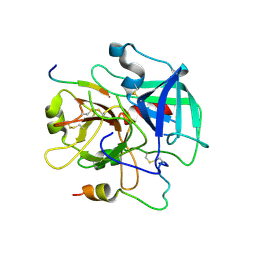 | | Crystal structure of the human thrombin mutant D102N in complex with the extracellular fragment of human PAR4. | | 分子名称: | Proteinase-activated receptor 4, Thrombin heavy chain, Thrombin light chain | | 著者 | Gandhi, P, Chen, Z, Appelbaum, E, Zapata, F, Di Cera, E. | | 登録日 | 2011-01-19 | | 公開日 | 2011-06-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of thrombin-protease-receptor interactions
IUBMB LIFE, 63, 2011
|
|
2M3K
 
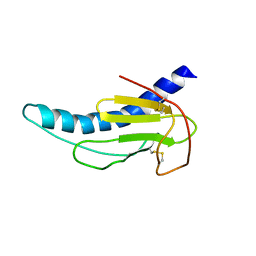 | |
7EXT
 
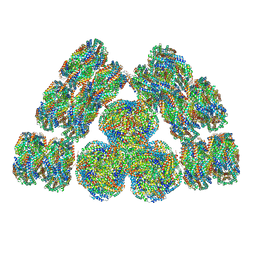 | | Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002 | | 分子名称: | Allophycocyanin alpha subunit, Allophycocyanin beta subunit, Allophycocyanin subunit alpha-B, ... | | 著者 | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | 登録日 | 2021-05-28 | | 公開日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
7EYD
 
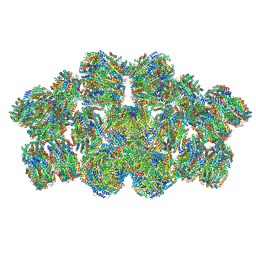 | | Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120 | | 分子名称: | Allophycocyanin subunit alpha 1, Allophycocyanin subunit alpha-B, Allophycocyanin subunit beta, ... | | 著者 | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | 登録日 | 2021-05-30 | | 公開日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
7NZJ
 
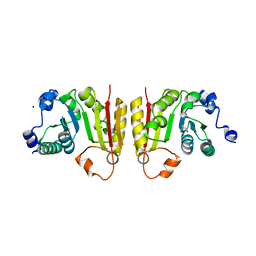 | | Structure of bsTrmB apo | | 分子名称: | GLYCEROL, SODIUM ION, tRNA (guanine-N(7)-)-methyltransferase | | 著者 | Blersch, K.F, Ficner, R, Neumann, P. | | 登録日 | 2021-03-24 | | 公開日 | 2021-09-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
5EB6
 
 | |
8I2G
 
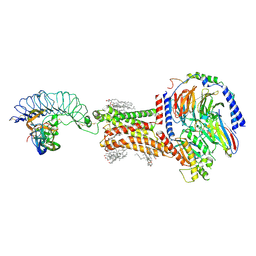 | | FSHR-Follicle stimulating hormone-compound 716340-Gs complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | 登録日 | 2023-01-14 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
3QL3
 
 | | Re-refined coordinates for PDB entry 1RX2 | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | 登録日 | 2011-02-02 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
