5NW2
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-3,3-dimethyl-2-(oxetane-3-carboxamido)butanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 19) | | 分子名称: | (2~{S},4~{R})-1-[(2~{S})-3,3-dimethyl-2-(oxetan-3-ylcarbonylamino)butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | 著者 | Gadd, M.S, Soares, P, Ciulli, A. | | 登録日 | 2017-05-04 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
2Y3G
 
 | | Se-Met form of Cupriavidus metallidurans CH34 CnrXs | | 分子名称: | CHLORIDE ION, GLYCEROL, NICKEL AND COBALT RESISTANCE PROTEIN CNRR, ... | | 著者 | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | 登録日 | 2010-12-20 | | 公開日 | 2011-03-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
7W6M
 
 | | Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | 登録日 | 2021-12-02 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
1YEZ
 
 | | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30. | | 分子名称: | MM1357 | | 著者 | Rossi, P, Aramini, J.M, Swapna, G.V.T, Huang, Y.P, Xiao, R, Ho, C.K, Ma, L.C, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-12-29 | | 公開日 | 2005-02-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30.
To be Published
|
|
7W73
 
 | | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | 登録日 | 2021-12-03 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
4IDQ
 
 | | human atlastin-1 1-446, N440T, GDPAlF4- | | 分子名称: | Atlastin-1, GUANOSINE-5'-DIPHOSPHATE, HEXAETHYLENE GLYCOL, ... | | 著者 | Byrnes, L.J, Singh, A, Szeto, K, Benvin, N.M, O'Donnell, J.P, Zipfel, W.R, Sondermann, H. | | 登録日 | 2012-12-12 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Structural basis for conformational switching and GTP loading of the large G protein atlastin.
Embo J., 32, 2013
|
|
5NW4
 
 | | Human cytoplasmic dynein-1 bound to dynactin and an N-terminal construct of BICD2 | | 分子名称: | Arp1, Arp11, BICD2N, ... | | 著者 | Zhang, K, Foster, H.E, Carter, A.P. | | 登録日 | 2017-05-05 | | 公開日 | 2017-08-02 | | 最終更新日 | 2018-11-21 | | 実験手法 | ELECTRON MICROSCOPY (8.7 Å) | | 主引用文献 | Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated.
Cell, 169, 2017
|
|
5NZV
 
 | | The structure of the COPI coat linkage IV | | 分子名称: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (17.299999 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
1I25
 
 | |
4QIM
 
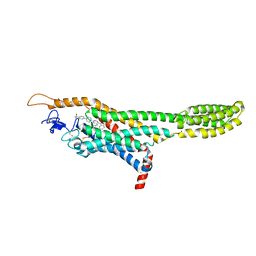 | | Structure of the human smoothened receptor in complex with ANTA XV | | 分子名称: | 2-{6-[4-(4-benzylphthalazin-1-yl)piperazin-1-yl]pyridin-3-yl}propan-2-ol, Smoothened homolog/Soluble cytochrome b562 chimeric protein, ZINC ION | | 著者 | Wang, C, Wu, H, Evron, T, Vardy, E, Han, G.W, Huang, X.-P, Hufeisen, S.J, Mangano, T.J, Urban, D.J, Katritch, V, Cherezov, V, Caron, M.G, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | 登録日 | 2014-05-31 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
4IDL
 
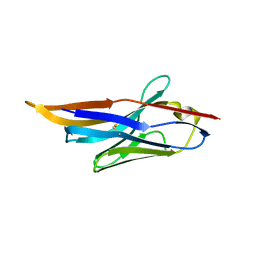 | |
4KZE
 
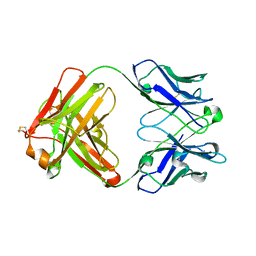 | | Crystal structure of an RNA aptamer in complex with Fab | | 分子名称: | BL3-6 Fab antibody, heavy chain, light chain, ... | | 著者 | Huang, H, Suslov, N.B, Li, N, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | 登録日 | 2013-05-29 | | 公開日 | 2014-06-18 | | 最終更新日 | 2014-07-30 | | 実験手法 | X-RAY DIFFRACTION (2.404 Å) | | 主引用文献 | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
7AZL
 
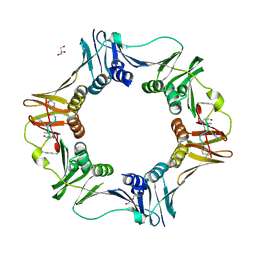 | | DNA polymerase sliding clamp from Escherichia coli with peptide 38 bound | | 分子名称: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ7
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 37 bound | | 分子名称: | Beta sliding clamp, FORMIC ACID, PENTAETHYLENE GLYCOL, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ6
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 36 bound | | 分子名称: | ACETATE ION, Beta sliding clamp, CHLORIDE ION, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
4C10
 
 | | Cryo-EM reconstruction of empty enterovirus 71 in complex with a neutralizing antibody E19 | | 分子名称: | CHLORIDE ION, EV19 5 C1-6 F1 C11, SODIUM ION, ... | | 著者 | Plevka, P, Perera, R, Cardosa, J, Suksatu, A, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2013-08-08 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (13 Å) | | 主引用文献 | Neutralizing Antibodies Can Initiate Genome Release from Human Enterovirus 71.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7K4Y
 
 | | Crystal structure of Kemp Eliminase HG3.17 at 343 K | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
2XZ9
 
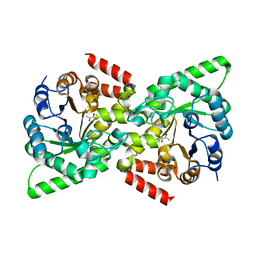 | | CRYSTAL STRUCTURE FROM THE PHOSPHOENOLPYRUVATE-BINDING DOMAIN OF ENZYME I IN COMPLEX WITH PYRUVATE FROM THE THERMOANAEROBACTER TENGCONGENSIS PEP-SUGAR PHOSPHOTRANSFERASE SYSTEM (PTS) | | 分子名称: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE-PROTEIN KINASE (PTS SYSTEM EI COMPONENT IN BACTERIA), PYRUVIC ACID | | 著者 | Oberholzer, A.E, Waltersperger, S.M, Schneider, P, Baumann, U, Erni, B. | | 登録日 | 2010-11-24 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.677 Å) | | 主引用文献 | Phosphoenolpyruvate: Sugar Phosphotransferase System from the Hyperthermophilic Thermoanaerobacter Tengcongensis.
Biochemistry, 50, 2011
|
|
3RUV
 
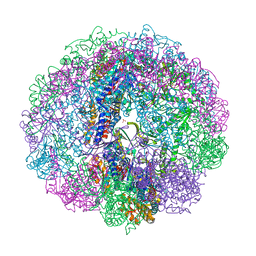 | | Crystal structure of Cpn-rls in complex with ATP analogue from Methanococcus maripaludis | | 分子名称: | Chaperonin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | 登録日 | 2011-05-05 | | 公開日 | 2012-01-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.242 Å) | | 主引用文献 | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
4QIN
 
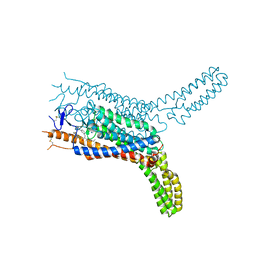 | | Structure of the human smoothened receptor in complex with SAG1.5 | | 分子名称: | 3-chloro-4,7-difluoro-N-[trans-4-(methylamino)cyclohexyl]-N-[3-(pyridin-4-yl)benzyl]-1-benzothiophene-2-carboxamide, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | 著者 | Wang, C, Wu, H, Evron, T, Vardy, E, Han, G.W, Huang, X.-P, Hufeisen, S.J, Mangano, T.J, Urban, D.J, Katritch, V, Cherezov, V, Caron, M.G, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | 登録日 | 2014-05-31 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
2CCI
 
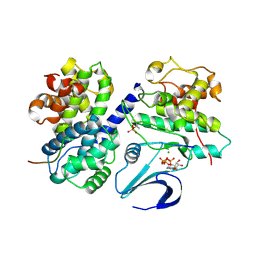 | | Crystal structure of phospho-CDK2 Cyclin A in complex with a peptide containing both the substrate and recruitment sites of CDC6 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6 homolog, Cyclin-A2, ... | | 著者 | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | 登録日 | 2006-01-16 | | 公開日 | 2006-05-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition.
J. Biol. Chem., 281, 2006
|
|
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | 著者 | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-30 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
7KCT
 
 | | Crystal Structure of the Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase (OGC) Biotin Carboxylase (BC) Domain Dimer in Complex with Adenosine 5'-Diphosphate Magnesium Salt (MgADP), Adenosine 5'-Diphosphate (ADP, and Bicarbonate Anion (Hydrogen Carbonate/HCO3-) | | 分子名称: | 2-oxoglutarate carboxylase small subunit, ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, ... | | 著者 | Buhrman, G.K, Rose, R.B, Enriquez, P, Truong, V. | | 登録日 | 2020-10-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structure, Function, and Thermal Adaptation of the Biotin Carboxylase Domain Dimer from Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase.
Biochemistry, 60, 2021
|
|
4BI9
 
 | | Crystal structure of wild-type SCP2 thiolase from Trypanosoma brucei. | | 分子名称: | 3-KETOACYL-COA THIOLASE, PUTATIVE | | 著者 | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | 登録日 | 2013-04-10 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
4BKQ
 
 | | Enoyl-ACP reductase from Yersinia pestis (wildtype)with cofactor NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPZ3_3519 | | 著者 | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | 登録日 | 2013-04-29 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
