8IRT
 
 | | Dopamine Receptor D3R-Gi-Rotigotine complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
8D9N
 
 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-06-10 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
6H07
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Nowotny, P, Biggel, P, Schneider, S, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Neutron and X-ray crystal structures of Lactobacillus brevis alcohol dehydrogenase reveal new insights into hydrogen-bonding pathways.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1PVP
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, ALSHG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | Descriptor: | 34-MER, Recombinase cre | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
5EBE
 
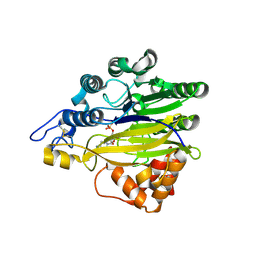 | | Structure of human sphingomyelinase phosphodiesterase like 3A (SMPDL3A) with 5' CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-O-phosphono-beta-D-ribofuranose, ... | | Authors: | Lim, S.M, Yeung, K, Tresaugues, L, Teo, H.L, Nordlund, P. | | Deposit date: | 2015-10-19 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid sphingomyelinase homologue with a novel nucleotide hydrolase activity.
Febs J., 283, 2016
|
|
8IRS
 
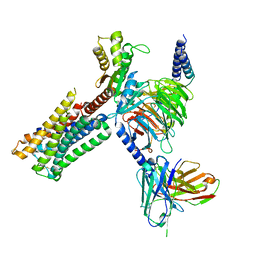 | | Dopamine Receptor D2R-Gi-Rotigotine complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
2VR7
 
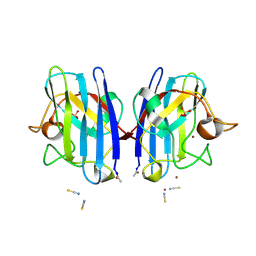 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.58 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
2JLN
 
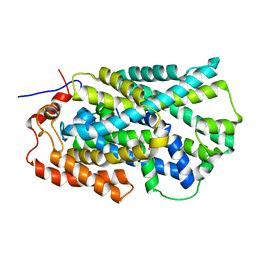 | | Structure of Mhp1, a nucleobase-cation-symport-1 family transporter | | Descriptor: | MERCURY (II) ION, MHP1, SODIUM ION | | Authors: | Weyand, S, Shimamura, T, Yajima, S, Suzuki, S, Mirza, O, Krusong, K, Carpenter, E.P, Rutherford, N.G, Hadden, J.M, O'Reilly, J, Ma, P, Saidijam, M, Patching, S.G, Hope, R.J, Norbertczak, H.T, Roach, P.C.J, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2008-09-11 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Molecular Mechanism of a Nucleobase-Cation-Symport-1 Family Transporter.
Science, 322, 2008
|
|
6MTJ
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-378806 in Complex with Human Antibodies 3H109L and 35O22 at 2.9 Angstrom | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-19 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
6X9F
 
 | | Pseudomonas aeruginosa MurC with AZ8074 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ8074
To Be Published
|
|
6SF7
 
 | | Atomic resolution structure of SplF protease from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Golik, P, Stach, N, Karim, A, Dubin, G. | | Deposit date: | 2019-08-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of Substrate Specificity of SplF Protease from Staphylococcus aureus .
Int J Mol Sci, 22, 2021
|
|
6XB4
 
 | |
7K8S
 
 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C002 (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C002 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Malyutin, A.G, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
8QCJ
 
 | |
4ZHM
 
 | |
7N6E
 
 | | TCR peptide HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1, ... | | Authors: | Chaurasia, P, Rossjohn, J, Petersen, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein.
J.Biol.Chem., 297, 2021
|
|
6N0Z
 
 | |
8I67
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 2,4-Thiazolidinedione, Form I | | Descriptor: | 1,2-ETHANEDIOL, 1,3-thiazolidine-2,4-dione, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8IRR
 
 | | Dopamine Receptor D1R-Gs-Rotigotine complex | | Descriptor: | CHOLESTEROL, D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
6N1Y
 
 | | Structure of L509V CAO1 - growth condition 1 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase, FE (II) ION | | Authors: | Khadka, N, Kiser, P.D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evidence for distinct rate-limiting steps in the cleavage of alkenes by carotenoid cleavage dioxygenases.
J.Biol.Chem., 294, 2019
|
|
2J8A
 
 | | X-ray structure of the N-terminus RRM domain of Set1 | | Descriptor: | HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC | | Authors: | Tresaugues, L, Dehe, P.M, Guerois, R, Rodriguez-Gil, A, Varlet, I, Salah, P, Pamblanco, M, Luciano, P, Quevillon-Cheruel, S, Sollier, J, Leulliot, N, Couprie, J, Tordera, V, Zinn-Justin, S, Chavez, S, Van Tilbeurgh, H, Geli, V. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structure of the N-Terminus Rrm Domain of Set1
To be Published
|
|
8I61
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I69
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Fluoroorotic acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
6XUC
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Chlorite dismutase | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8702 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
6N21
 
 | | Structure of wild-type CAO1 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase, FE (II) ION | | Authors: | Khadka, N, Kiser, P.D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Evidence for distinct rate-limiting steps in the cleavage of alkenes by carotenoid cleavage dioxygenases.
J.Biol.Chem., 294, 2019
|
|
