1ZF5
 
 | | GCT duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*CP*GP*CP*TP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7WQI
 
 | | Crystal structure of SARS coronavirus main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of SARS coronavirus main protease in complex with PF07304814
To Be Published
|
|
7JM1
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Aminocyclitol acetyltransferase ApmA | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA
To Be Published
|
|
4EAA
 
 | | X-ray crystal structure of the H141N mutant of perosamine N-acetyltransferase from Caulobacter crescentus in complex with CoA and GDP-perosamine | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-perosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
1FFW
 
 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY WITH A BOUND IMIDO DIPHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, IMIDO DIPHOSPHATE, ... | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7K4U
 
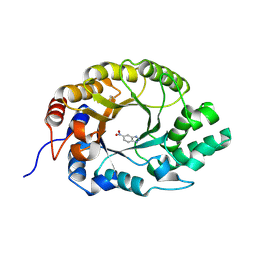 | | Crystal structure of Kemp Eliminase HG3 K50Q in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
5NQA
 
 | | Crystal structure of GalNAc-T4 in complex with the monoglycopeptide 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, GLYCEROL, ... | | Authors: | de las Rivas, M, Lira-Navarrete, E, Daniel, E.J.P, Companon, I, Coelho, H, Diniz, A, Jimenez-Barbero, J, Peregrina, J.M, Clausen, H, Corzana, F, Marcelo, F, Jimenez-Oses, G, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The interdomain flexible linker of the polypeptide GalNAc transferases dictates their long-range glycosylation preferences.
Nat Commun, 8, 2017
|
|
1ZA2
 
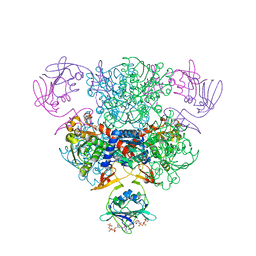 | | Structure of wild-type E. coli Aspartate Transcarbamoylase in the presence of CTP, carbamoyl phosphate at 2.50 A resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, J, Stieglitz, K.A, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7KDG
 
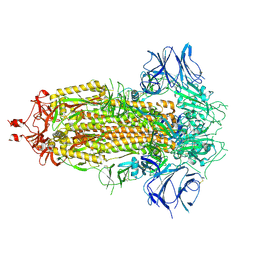 | |
7LOL
 
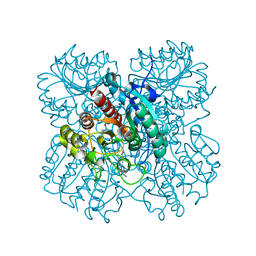 | | The structure of Agmatinase from E. Coli at 1.8 A displaying urea and agmatine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGMATINE, Agmatinase, ... | | Authors: | Maturana, P, Figueroa, M, Gonzalez-Ordenes, F, Villalobos, P, Martinez-Oyanedel, J, Uribe, E.A, Castro-Fernandez, V. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity.
Int J Mol Sci, 22, 2021
|
|
2MTZ
 
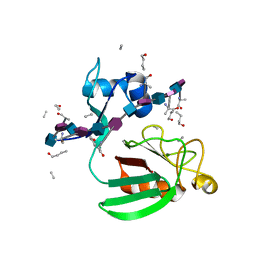 | | Haddock model of Bacillus subtilis L,D-transpeptidase in complex with a peptidoglycan hexamuropeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Putative L,D-transpeptidase YkuD, intact bacterial peptidoglycan | | Authors: | Schanda, P, Triboulet, S, Laguri, C, Bougault, C, Ayala, I, Callon, M, Arthur, M, Simorre, J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan.
J.Am.Chem.Soc., 136, 2014
|
|
1K42
 
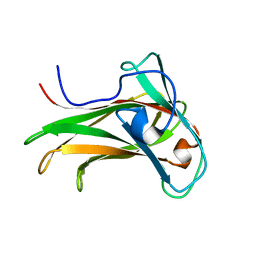 | | The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. | | Descriptor: | Xylanase | | Authors: | Simpson, P.J, Jamieson, S.J, Abou-Hachem, M, Nordberg-Karlsson, E, Gilbert, H.J, Holst, O, Williamson, M.P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase.
Biochemistry, 41, 2002
|
|
1FE8
 
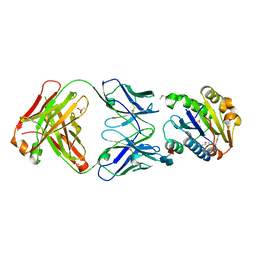 | | CRYSTAL STRUCTURE OF THE VON WILLEBRAND FACTOR A3 DOMAIN IN COMPLEX WITH A FAB FRAGMENT OF IGG RU5 THAT INHIBITS COLLAGEN BINDING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, IMMUNOGLOBULIN IGG RU5, ... | | Authors: | Bouma, B, Huizinga, E.G, Schiphorst, M.E, Sixma, J.J, Kroon, J, Gros, P. | | Deposit date: | 2000-07-21 | | Release date: | 2001-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of the collagen-binding site of the von Willebrand factor A3-domain.
J.Biol.Chem., 276, 2001
|
|
1P69
 
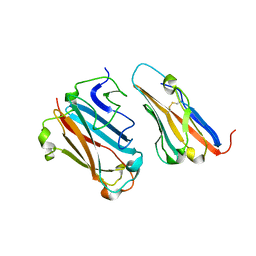 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (P417S MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
4PK4
 
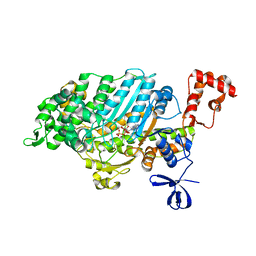 | | Myosin VI motor domain in the PPS state - from a Pi release state crystal, space group P212121 after long soaking with PO4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Isabet, T, Benisty, H, Llinas, P, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2014-05-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | How actin initiates the motor activity of Myosin.
Dev.Cell, 33, 2015
|
|
4LM6
 
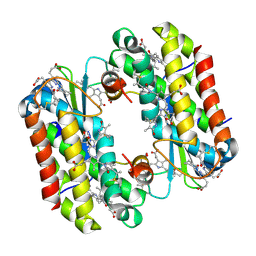 | | Light harvesting complex PC612 from the cryptophyte Hemiselmis virescens M1635 | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, cryptophyte phycocyanin alpha chain, ... | | Authors: | Harrop, S.J, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2013-07-10 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Single-residue insertion switches the quaternary structure and exciton states of cryptophyte light-harvesting proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2N4S
 
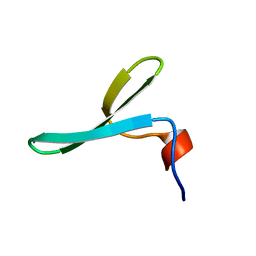 | |
5NVV
 
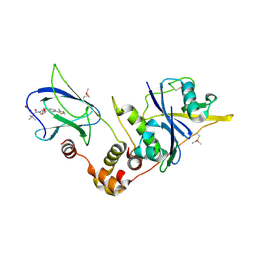 | | pVHL:EloB:EloC in complex with (2S,4R)-4-hydroxy-1-((S)-2-(2-hydroxyacetamido)-3,3-dimethylbutanoyl)-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-3,3-dimethyl-2-(2-oxidanylethanoylamino)butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
4PMO
 
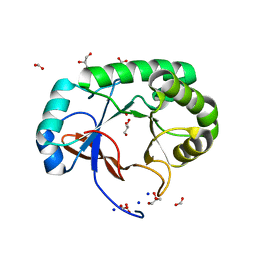 | | Crystal structure of the Mycobacterium tuberculosis Tat-secreted protein Rv2525c, monoclinic crystal form I | | Descriptor: | FORMIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Bellinzoni, M, Haouz, A, Shepard, W, Alzari, P.M. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural studies suggest a peptidoglycan hydrolase function for the Mycobacterium tuberculosis Tat-secreted protein Rv2525c.
J.Struct.Biol., 188, 2014
|
|
4LMD
 
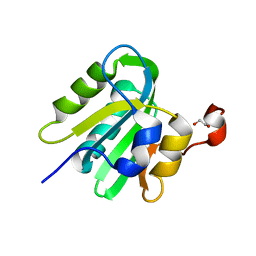 | |
1R4S
 
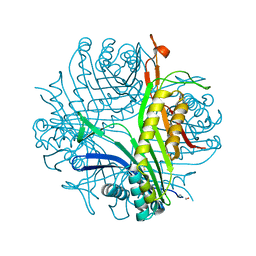 | | URATE OXIDASE FROM ASPERGILLUS FLAVUS COMPLEXED WITH ITS INHIBITOR 9-METHYL URIC ACID | | Descriptor: | 9-METHYL URIC ACID, CYSTEINE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Prange, T. | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7LJM
 
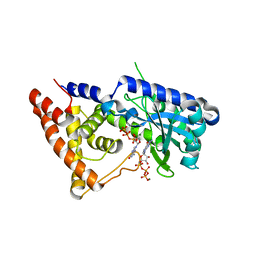 | | Structure of the Salmonella enterica CD-NTase CdnD in complex with GTP | | Descriptor: | CD-NTase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.W, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
1R54
 
 | | Crystal structure of the catalytic domain of human ADAM33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADAM 33, CALCIUM ION, ... | | Authors: | Orth, P, Reicher, P, Wang, W, Prosise, W.W, Yarosh-Tomaine, T, Hammond, G, Xiao, L, Mirza, U.A, Zou, J, Strickland, C, Taremi, S.S. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structre of the catalytic domain of human ADAM33
J.Mol.Biol., 335, 2004
|
|
4LN4
 
 | | The crystal structure of hemagglutinin form a h7n9 influenza virus (a/shanghai/1/2013) in complex with lstb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Analysis of the Hemagglutinin from the Recent 2013 H7N9 Influenza Virus.
J.Virol., 87, 2013
|
|
7LJO
 
 | | Structure of the Bacteroides fragilis CD-NTase CdnB in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CD-NTase, MAGNESIUM ION | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.T, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
