2J8J
 
 | | Solution Structure of the A4 Domain of Blood Coagulation Factor XI | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4E36
 
 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 variant N392K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Birtley, J.R, Saridakis, E, Pegias, P, Stratikos, E, Mavridis, I.M. | | Deposit date: | 2012-03-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A common single nucleotide polymorphism in endoplasmic reticulum aminopeptidase 2 induces a specificity switch that leads to altered antigen processing.
J.Immunol., 189, 2012
|
|
4Q6X
 
 | | Structure of phospholipase D Beta1B1i from Sicarius terrosus venom at 2.14 A resolution | | Descriptor: | MAGNESIUM ION, Phospholipase D StSicTox-betaIC1 | | Authors: | Lajoie, D.M, Roberts, S.A, Zobel-Thropp, P.A, Binford, G.J, Cordes, M.H. | | Deposit date: | 2014-04-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Variable Substrate Preference among Phospholipase D Toxins from Sicariid Spiders.
J.Biol.Chem., 290, 2015
|
|
1XI6
 
 | | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001 | | Descriptor: | extragenic suppressor | | Authors: | Zhao, M, Chang, J.C, Zhou, W, Chen, L, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001
To be published
|
|
2ZZ8
 
 | |
1S0P
 
 | | Structure of the N-Terminal Domain of the Adenylyl Cyclase-Associated Protein (CAP) from Dictyostelium discoideum. | | Descriptor: | Adenylyl cyclase-associated protein, MAGNESIUM ION | | Authors: | Ksiazek, D, Brandstetter, H, Israel, L, Bourenkov, G.P, Katchalova, G, Janssen, K.P, Bartunik, H.D, Noegel, A.A, Schleicher, M, Holak, T.A. | | Deposit date: | 2004-01-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE ADENYLYL
CYCLASE-ASSOCIATED PROTEIN (CAP) FROM DICTYOSTELIUM DISCOIDEUM
Structure, 11, 2003
|
|
5I67
 
 | | Crystal Structure Analysis of MTB PEPCK mutant C273S | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Dostal, J, Pachl, P, Machova, I, Pichova, I, Snasel, J. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal Structure Analysis of MTB PEPCK mutant C273S
To Be Published
|
|
1XJK
 
 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dGTP-ADP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3IF4
 
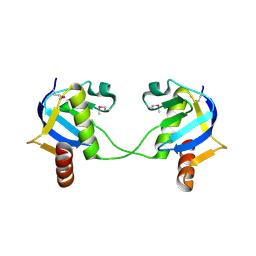 | | Structure from the mobile metagenome of North West Arm Sewage Outfall: Integron Cassette Protein Hfx_Cass5 | | Descriptor: | Integron Cassette Protein Hfx_Cass5 | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Evdokimova, E, Kudrytska, M, Koenig, J.E, Osipiuk, J, Edwards, A, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-24 | | Release date: | 2009-09-08 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites.
Plos One, 8, 2013
|
|
5I7E
 
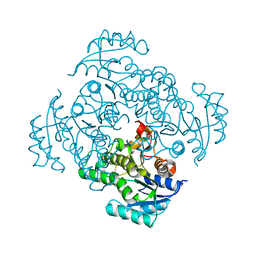 | |
7WSJ
 
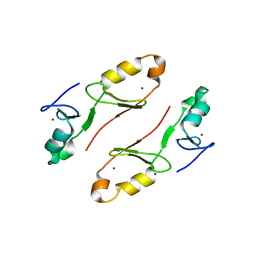 | | Crystal structure of the tandem B-box domain of Arabidopsis thaliana CONSTANS | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS | | Authors: | Dahal, P, Pathak, D, Kwon, E, Kim, D.Y. | | Deposit date: | 2022-01-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a tandem B-box domain from Arabidopsis CONSTANS.
Biochem.Biophys.Res.Commun., 599, 2022
|
|
7KI0
 
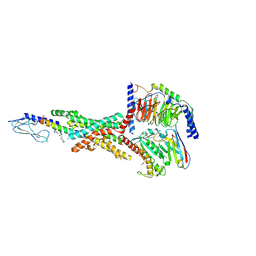 | | Semaglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs protein | | Descriptor: | 17-amino-10-oxo-3,6,12,15-tetraoxa-9-azaheptadecan-1-oic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Belousoff, M.J, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2020-10-22 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and dynamics of semaglutide- and taspoglutide-bound GLP-1R-Gs complexes.
Cell Rep, 36, 2021
|
|
4LAU
 
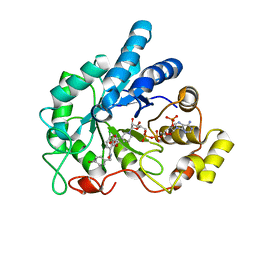 | | Crystal structure of human AR complexed with NADP+ and {2-[(4-bromobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-bromobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-20 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.843 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
7KI1
 
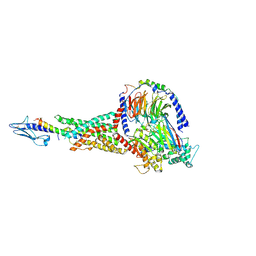 | | Taspoglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs Protein | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, X, Belousoff, M.J, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2020-10-22 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and dynamics of semaglutide- and taspoglutide-bound GLP-1R-Gs complexes.
Cell Rep, 36, 2021
|
|
1P9U
 
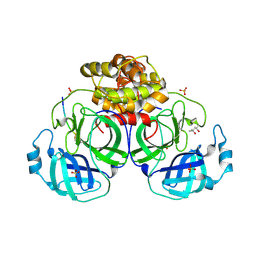 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHQ-VNSTLQ-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION, ... | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
1DIP
 
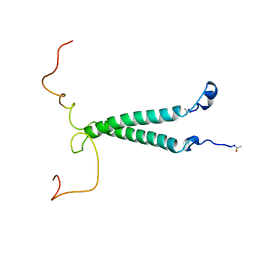 | | THE SOLUTION STRUCTURE OF PORCINE DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE, NMR, 10 STRUCTURES | | Descriptor: | DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE | | Authors: | Roesch, P, Seidel, G, Adermann, K, Schindler, T, Ejchart, A, Jaenicke, R, Forssmann, W.G. | | Deposit date: | 1997-04-09 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine delta sleep-inducing peptide immunoreactive peptide A homolog of the shortsighted gene product.
J.Biol.Chem., 272, 1997
|
|
1XOS
 
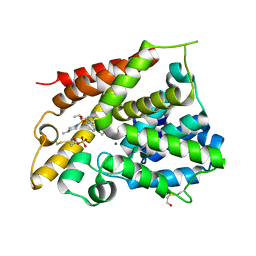 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Sildenafil | | Descriptor: | 5-{2-ETHOXY-5-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-1-METHYL-3-PROPYL-1H,6H,7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
2DQY
 
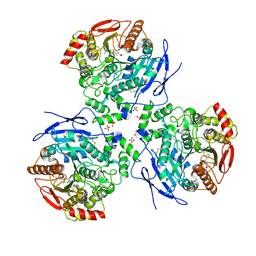 | | Crystal structure of human carboxylesterase in complex with cholate and palmitate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLIC ACID, Liver carboxylesterase 1, ... | | Authors: | Bencharit, S, Edwards, C.C, Morton, C.L, Howard-Williams, E.L, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1
J.Mol.Biol., 363, 2006
|
|
1XQM
 
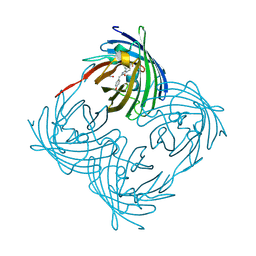 | | Variations on the GFP chromophore scaffold: A fragmented 5-membered heterocycle revealed in the 2.1A crystal structure of a non-fluorescent chromoprotein | | Descriptor: | ACETIC ACID, kindling fluorescent protein | | Authors: | Wilmann, P.G, Petersen, J, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variations on the GFP chromophore: A polypeptide fragmentation within the chromophore revealed in the 2.1-A crystal structure of a nonfluorescent chromoprotein from Anemonia sulcata
J.Biol.Chem., 280, 2005
|
|
1XOZ
 
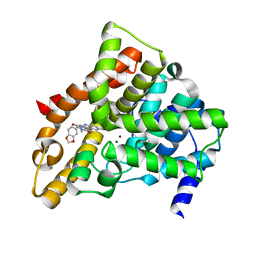 | | Catalytic Domain Of Human Phosphodiesterase 5A In Complex With Tadalafil | | Descriptor: | 6-BENZO[1,3]DIOXOL-5-YL-2-METHYL-2,3,6,7,12,12A-HEXAHYDRO-PYRAZINO[1',2':1,6]PYRIDO[3,4-B]INDOLE-1,4-DIONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-07 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
1XRF
 
 | | The Crystal Structure of a Novel, Latent Dihydroorotase from Aquifex aeolicus at 1.7 A resolution | | Descriptor: | Dihydroorotase, SULFATE ION, ZINC ION | | Authors: | Martin, P.D, Purcarea, C, Zhang, P, Vaishnav, A, Sadecki, S, Guy-Evans, H.I, Evans, D.R, Edwards, B.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a novel, latent dihydroorotase from Aquifex aeolicus at 1.7A resolution
J.Mol.Biol., 348, 2005
|
|
4LHO
 
 | | Crystal Structure of FG41Malonate Semialdehyde Decarboxylase inhibited by 3-bromopropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
1JWT
 
 | | CRYSTAL STRUCTURE OF THROMBIN IN COMPLEX WITH A NOVEL BICYCLIC LACTAM INHIBITOR | | Descriptor: | 4-OXO-2-PHENYLMETHANESULFONYL-OCTAHYDRO-PYRROLO[1,2-A]PYRAZINE-6-CARBOXYLIC ACID [1-(N-HYDROXYCARBAMIMIDOYL)-PIPERIDIN-4-YLMETHYL]-AMIDE, Prothrombin | | Authors: | Levesque, S, St-Denis, Y, Bachand, B, Preville, P, Leblond, L, Winocour, P.D, Edmunds, J.J, Rubin, J.R, Siddiqui, M.A. | | Deposit date: | 2001-09-05 | | Release date: | 2002-02-27 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel bicyclic lactam inhibitors of thrombin: potency and selectivity optimization through P1 residues.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
2M27
 
 | | Major G-quadruplex structure formed in human VEGF promoter, a monomeric parallel-stranded quadruplex | | Descriptor: | DNA_(5'-D(*CP*GP*GP*GP*GP*CP*GP*GP*GP*CP*CP*TP*TP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3')_ | | Authors: | Agrawal, P, Hatzakis, E, Guo, K, Carver, M, Yang, D. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes.
Nucleic Acids Res., 41, 2013
|
|
3I7L
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of DDB2 | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
