9FY4
 
 | | Crystal structure of Heme-Oxygenase mutant I143K from Corynebacterium diphtheriae complexed with Cobalt-porphyrine (HumO-Co(III)) | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Labidi, R.J, Faivre, B, Carpentier, P, Perard, J, Gotico, P, Li, Y, Atta, M, Fontecave, M. | | Deposit date: | 2024-07-02 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | Light-Activated Artificial CO 2 -Reductase: Structure and Activity.
J.Am.Chem.Soc., 2024
|
|
7T7M
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
3LT4
 
 | | Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan variant PB4 | | Descriptor: | 2-(2-amino-4-chlorophenoxy)-5-chlorophenol, Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Maity, K, Bhargav, S.P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystallographic analysis of the complexes of enoyl acyl carrier protein reductase of Plasmodium falciparum with triclosan variants to elucidate the importance of different functional groups in enzyme inhibition
Iubmb Life, 62, 2010
|
|
6DBY
 
 | | Crystal structure of Nudix 1 from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, Nudix hydrolase 1 | | Authors: | Noel, J.P, Thomas, S.T, Dudareva, N, Henry, L.K. | | Deposit date: | 2018-05-03 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of isopentenyl phosphate to plant terpenoid metabolism.
Nat Plants, 4, 2018
|
|
9ES8
 
 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f with decylplastoquinone bound at plastoquionol reduction site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
6DC0
 
 | |
6QG4
 
 | |
6DR5
 
 | |
9FP0
 
 | | Cryo-EM structure of the 'crown'less Bcs macrocomplex for E. coli cellulose secretion in non-saturating c-di-GMP (local) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ADENOSINE-5'-TRIPHOSPHATE, Cell division protein, ... | | Authors: | Anso, I, Krasteva, P.V. | | Deposit date: | 2024-06-12 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system.
Nat Commun, 15, 2024
|
|
9GQO
 
 | | Structure of a Ca2+ bound phosphoenzyme intermediate in the inward-to-outward transition of Ca2+-ATPase 1 from Listeria monocytogenes | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase, MAGNESIUM ION, ... | | Authors: | Hansen, S.B, Flygaard, R.F, Kjaergaard, M, Nissen, P. | | Deposit date: | 2024-09-09 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of a Ca2+ bound phosphoenzyme intermediate in the inward-to-outward transition of Ca2+-ATPase 1 from Listeria monocytogenes
To Be Published
|
|
3L8K
 
 | | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dihydrolipoyl dehydrogenase, PHOSPHATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus
To be Published
|
|
9ES7
 
 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with water molecules at 1.94 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
6Q2Y
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
3LAU
 
 | |
9DEQ
 
 | | Cryo-EM structures of full-length integrin alphaIIbbeta3 in native lipids complexed with modified tirofiban | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adair, B, Xiong, J.P, Yeager, M, Arnaout, M.A. | | Deposit date: | 2024-08-29 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Platelet integrin alpha IIb beta 3 plays a key role in a venous thrombogenesis mouse model.
Nat Commun, 15, 2024
|
|
4WS6
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-aminouracil, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7BOI
 
 | | Bacterial 30S ribosomal subunit assembly complex state F (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7BOF
 
 | | Bacterial 30S ribosomal subunit assembly complex state I (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
6Q60
 
 | | Structure of GluA2 ligand-binding domain (S1S2J) in complex with the agonist (S)-2-Amino-3-(2-methyl-5-hydroxy-2H-1,2,3-triazol-4-yl)propanoic acid at 1.55 A resolution | | Descriptor: | (2~{S})-2-azanyl-3-(2-methyl-5-oxidanyl-1,2,3-triazol-4-yl)propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Moellerud, S, Temperini, P, Kastrup, J.S. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Use of the 4-Hydroxytriazole Moiety as a Bioisosteric Tool in the Development of Ionotropic Glutamate Receptor Ligands.
J.Med.Chem., 62, 2019
|
|
7BOD
 
 | | Bacterial 30S ribosomal subunit assembly complex state M (body domain) | | Descriptor: | 16S rRNA (body domain of 30S subunit), 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S.R. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
8SVY
 
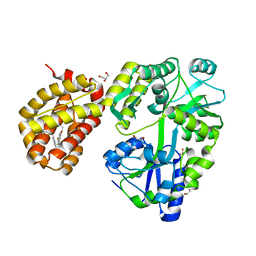 | | MBP-Mcl1 in complex with ligand 10 | | Descriptor: | (15P)-17-chloro-33-fluoro-12-[(2-methoxyethoxy)methyl]-5,14,22-trimethyl-28-oxa-9-thia-5,6,13,14,22-pentaazaheptacyclo[27.7.1.1~4,7~.0~11,15~.0~16,21~.0~20,24~.0~30,35~]octatriaconta-1(36),4(38),6,11(15),12,16,18,20,23,29(37),30,32,34-tridecaene-23-carboxylic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Macrocyclic Carbon-Linked Pyrazoles As Novel Inhibitors of MCL-1.
Acs Med.Chem.Lett., 14, 2023
|
|
3LFN
 
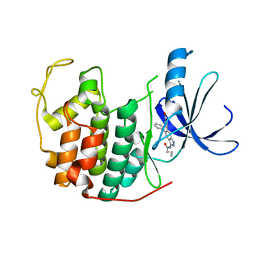 | | Crystal structure of CDK2 with SAR57, an aminoindazole type inhibitor | | Descriptor: | Cell division protein kinase 2, N-[6-(4-hydroxyphenyl)-5-phenyl-1H-indazol-3-yl]butanamide | | Authors: | Dreyer, M.K, Wendt, K.U, Schimanski-Breves, S, Loenze, P. | | Deposit date: | 2010-01-18 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Rational design of potent GSK3beta inhibitors with selectivity for Cdk1 and Cdk2.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6D9T
 
 | | BshA from Staphylococcus aureus complexed with UDP | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Royer, C.R, Cook, P.D. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and functional analysis of the glycosyltransferase BshA from Staphylococcus aureus: Insights into the reaction mechanism and regulation of bacillithiol production.
Protein Sci., 28, 2019
|
|
7BOG
 
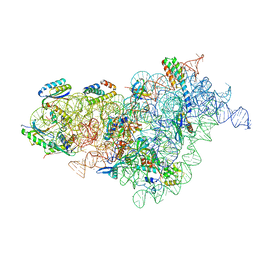 | | Bacterial 30S ribosomal subunit assembly complex state E (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
3LEO
 
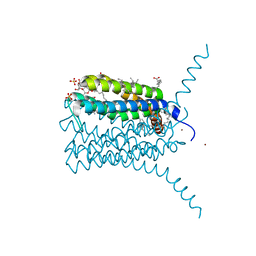 | | Structure of human Leukotriene C4 synthase mutant R31Q in complex with glutathione | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Niegowski, D, Martinez-Molina, D, Rinaldo-Matthis, A, Nordlund, P, Haeggstrom, J. | | Deposit date: | 2010-01-15 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arginine 104 is a key catalytic residue in leukotriene C4 synthase.
J.Biol.Chem., 285, 2010
|
|
