4BWB
 
 | | Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN, NEUROTENSIN RECEPTOR TYPE 1 | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-07-01 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2LTJ
 
 | | Conformational analysis of StrH, the surface-attached exo- beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Pluvinage, B, Chitayat, S, Ficko-Blean, E, Abbott, D, Kunjachen, J, Grondin, J, Spencer, H, Smith, S, Boraston, A. | | Deposit date: | 2012-05-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational analysis of StrH, the surface-attached exo-beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae.
J.Mol.Biol., 425, 2013
|
|
4A93
 
 | | RNA Polymerase II elongation complex containing a CPD Lesion | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*T*TTP*TP*TP*CP*C BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP*AP)-3', ... | | Authors: | Walmacq, C, Cheung, A.C.M, Kireeva, M.L, Lubkowska, L, Ye, C, Gotte, D, Strathern, J.N, Carell, T, Cramer, P, Kashlev, M. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Translesion Transcription by RNA Polymerase II and its Role in Cellular Resistance to DNA Damage.
Mol.Cell, 46, 2012
|
|
5CX8
 
 | | Structure of RagB, a major immunodominant virulence factor of Porphyromonas gingivalis. | | Descriptor: | 3-deoxy-5-O-phosphono-beta-D-ribofuranose, 3-deoxy-beta-D-glucopyranose, 6-O-phosphono-D-tagatose, ... | | Authors: | Goulas, T, Garcia-Ferrer, I, Hutcherson, J.A, Potempa, B.A, Potempa, J, Scott, D.A, Gomis-Ruth, F.X. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of RagB, a major immunodominant outer-membrane surface receptor antigen of Porphyromonas gingivalis.
Mol Oral Microbiol, 31, 2016
|
|
3NEW
 
 | | p38-alpha complexed with Compound 10 | | Descriptor: | 4-(trifluoromethyl)-3-[3-(trifluoromethyl)phenyl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Goedken, E.R, Comess, K.M, Sun, C, Argiriadi, M, Jia, Y, Quinn, C.M, Banach, D.L, Marcotte, D, Borhani, D. | | Deposit date: | 2010-06-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Characterization of Non-ATP Site Inhibitors of the Mitogen Activated Protein (MAP) Kinases.
Acs Chem.Biol., 6, 2011
|
|
4IN4
 
 | | Crystal structure of cpd 15 bound to Keap1 Kelch domain | | Descriptor: | 2-({5-[(2,4-dimethylphenyl)sulfonyl]-6-oxo-1,6-dihydropyrimidin-2-yl}sulfanyl)-N-[2-(trifluoromethyl)phenyl]acetamide, Kelch-like ECH-associated protein 1, PHOSPHATE ION | | Authors: | Silvian, L, Marcotte, D. | | Deposit date: | 2013-01-03 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism.
Bioorg.Med.Chem., 21, 2013
|
|
2VNO
 
 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens in complex with blood group B-trisaccharide ligand. | | Descriptor: | CALCIUM ION, CPE0329, alpha-L-fucopyranose-(1-2)-[beta-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-02-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
3R21
 
 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RTR
 
 | | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Calabrese, M.F, Scott, D.C, Duda, D.M, Grace, C.R, Kurinov, I, Kriwacki, R.W, Schulman, B.A. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1UPF
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V BOUND TO THE DRUG 5-FLUOROURACIL | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1BMP
 
 | | BONE MORPHOGENETIC PROTEIN-7 | | Descriptor: | BONE MORPHOGENETIC PROTEIN-7 | | Authors: | Griffith, D.L, Scott, D.L. | | Deposit date: | 1995-12-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1POB
 
 | | CRYSTAL STRUCTURE OF COBRA-VENOM PHOSPHOLIPASE A2 IN A COMPLEX WITH A TRANSITION-STATE ANALOGUE | | Descriptor: | 1-O-OCTYL-2-HEPTYLPHOSPHONYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | White, S.P, Scott, D.L, Otwinowski, Z, Sigler, P.B. | | Deposit date: | 1992-09-07 | | Release date: | 1993-10-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of cobra-venom phospholipase A2 in a complex with a transition-state analogue.
Science, 250, 1990
|
|
1DY3
 
 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
1UPU
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V, BOUND TO PRODUCT URIDINE-1-MONOPHOSPHATE (UMP) | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-04-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
2YL5
 
 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, MAGNESIUM ION | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
3R22
 
 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesana, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1BD3
 
 | | STRUCTURE OF THE APO URACIL PHOSPHORIBOSYLTRANSFERASE, 2 MUTANT C128V | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-05-12 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
2W7Y
 
 | | Structure of a Streptococcus pneumoniae solute-binding protein in complex with the blood group A-trisaccharide. | | Descriptor: | IODIDE ION, PROBABLE SUGAR ABC TRANSPORTER, SUGAR-BINDING PROTEIN, ... | | Authors: | Higgins, M.A, Abbott, D.W, Boulanger, M.J, Boraston, A.B. | | Deposit date: | 2009-01-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Blood-Group Antigen Recognition by a Solute-Binding Protein from a Serotype 3 Strain of Streptococcus Pneumoniae.
J.Mol.Biol., 388, 2009
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
1BD4
 
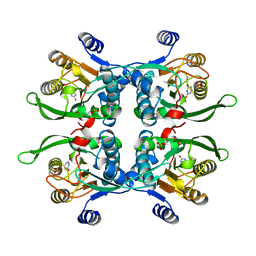 | | UPRT-URACIL COMPLEX | | Descriptor: | PHOSPHATE ION, URACIL, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-05-12 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
4DHF
 
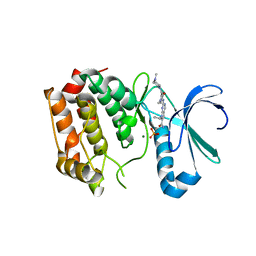 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4CSS
 
 | | Crystal structure of FimH in complex with a sulfonamide biphenyl alpha D-mannoside | | Descriptor: | 4'-(alpha-D-Mannopyranosyloxy)-biphenyl-4-methyl sulfonamide, GLYCEROL, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
2YL8
 
 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
2YL9
 
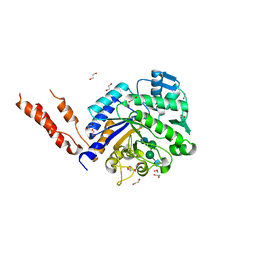 | | INHIBITION OF THE PNEUMOCOCCAL VIRULENCE FACTOR STRH AND MOLECULAR INSIGHTS INTO N-GLYCAN RECOGNITION AND HYDROLYSIS | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-06-01 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
1JV2
 
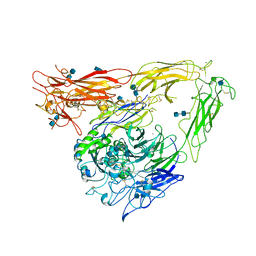 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHAVBETA3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.P, Stehle, T, Diefenbach, B, Zhang, R, Dunker, R, Scott, D, Joachimiak, A, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3.
Science, 294, 2001
|
|
