3VTC
 
 | | Crystal structure of rat vitamin D receptor bound to a partial agonist 26-adamantyl-23-yne-19-norvitammin D ADTK3 | | Descriptor: | (1R,3R,7E,17beta)-17-{(2R,6R)-6-hydroxy-7-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hept-4-yn-2-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 1,2-ETHANEDIOL, COACTIVATOR PEPTIDE DRIP, ... | | Authors: | Nakabayashi, M, Kudo, T, Tokiwa, H, Makishima, M, Yamada, S, Ikura, T, Ito, N. | | Deposit date: | 2012-05-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combination of Triple Bond and Adamantane Ring on the Vitamin D Side Chain Produced Partial Agonists for Vitamin D Receptor.
J.Med.Chem., 2014
|
|
3VTB
 
 | | Crystal structure of rat vitamin D receptor bound to a partial agonist 25-adamantyl-23-yne-19-norvitammin D ADTK1 | | Descriptor: | (1R,3R,7E,17beta)-17-{(2R,6S)-6-hydroxy-6-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hex-4-yn-2-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Kudo, T, Tokiwa, H, Makishima, M, Yamada, S, Ikura, T, Ito, N. | | Deposit date: | 2012-05-26 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combination of Triple Bond and Adamantane Ring on the Vitamin D Side Chain Produced Partial Agonists for Vitamin D Receptor.
J.Med.Chem., 2014
|
|
4Y7P
 
 | | Structure of alkaline D-peptidase from Bacillus cereus | | Descriptor: | Alkaline D-peptidase, THIOCYANATE ION | | Authors: | Nakano, S, Okazaki, S, Ishitsubo, E, Kawahara, N, Komeda, H, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-02-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational analysis of peptide recognition mechanism of class-C type penicillin binding protein, alkaline D-peptidase from Bacillus cereus DF4-B
Sci Rep, 5, 2015
|
|
4TRP
 
 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen | | Authors: | Horwacik, I, Golik, P, Grudnik, P, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUL
 
 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | ACETATE ION, Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, ... | | Authors: | Golik, P, Grudnik, P, Dubin, G, Zdzalik, M, Rokita, H, Horwacik, I. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUJ
 
 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, peptide1 | | Authors: | Grudnik, P, Golik, P, Horwacik, I, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUK
 
 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | CHLORIDE ION, Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, ... | | Authors: | Golik, P, Grudnik, P, Dubin, G, Horwacik, I, Zdzalik, M, Rokita, H. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUO
 
 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-[2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Golik, P, Grudnik, P, Horwacik, I, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
6JNO
 
 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
6JNR
 
 | | RXRa structure complexed with CU-6PMN and SRC1 peptide. | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RXRa structure complexed with CU-6PMN and SRC1 peptide.
To Be Published
|
|
4YJF
 
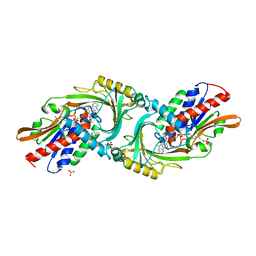 | | Crystal structure of DAAO(Y228L/R283G) variant (S-methylbenzylamine binding form) | | Descriptor: | (1S)-1-phenylethanamine, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakano, S, Yasukawa, K, Kawahara, N, Ishitsubo, E, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DAAO variant
To Be Published
|
|
4YJD
 
 | | Crystal structure of DAAO(Y228L/R283G) variant (apo form) | | Descriptor: | D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Nakano, S, Yasukawa, K, Kawahara, N, Ishitsubo, E, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of DAAO variant
To Be Published
|
|
4YJG
 
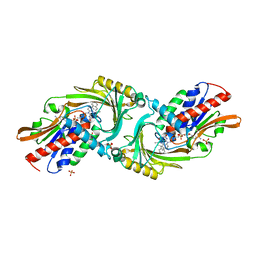 | | Crystal structure of DAAO(Y228L/R283G) variant (R-3-amino 1-phenylbutane binding form) | | Descriptor: | (2R)-4-phenylbutan-2-amine, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakano, S, Yasukawa, K, Kawahara, N, Ishitsubo, E, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DAAO variant
To Be Published
|
|
4YJH
 
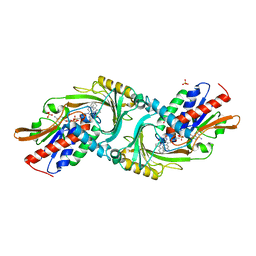 | | Crystal structure of DAAO(Y228L/R283G) variant (R-2-phenylpyrrolidine binding form) | | Descriptor: | (2R)-2-phenylpyrrolidine, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakano, S, Yasukawa, K, Kawahara, N, Ishitsubo, E, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DAAO variant
To Be Published
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
6L96
 
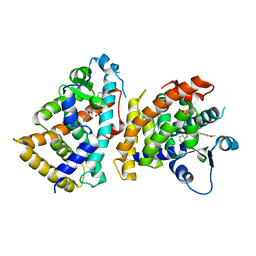 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | Authors: | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
5ZQU
 
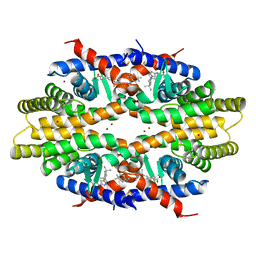 | | Crystal structure of tetrameric RXRalpha-LBD complexed with partial agonist CBt-PMN | | Descriptor: | 1-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)benzotriazole-5-carboxylic acid, BROMIDE ION, Retinoic acid receptor RXR-alpha | | Authors: | Miyashita, Y, Numoto, N, Arulmozhiraja, S, Nakano, S, Matsuo, N, Shimizu, K, Kakuta, H, Ito, S, Ikura, T, Ito, N, Tokiwa, H. | | Deposit date: | 2018-04-20 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.60038781 Å) | | Cite: | Dual conformation of the ligand induces the partial agonistic activity of retinoid X receptor alpha (RXR alpha ).
FEBS Lett., 593, 2019
|
|
3VTD
 
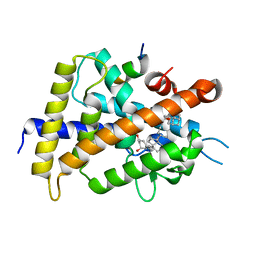 | | Crystal structure of rat vitamin D receptor bound to a partial agonist 26-adamantyl-23-yne-19-norvitammin D ADTK4 | | Descriptor: | (1R,3R,7E,17beta)-17-{(2R,6S)-6-hydroxy-7-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hept-4-yn-2-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Kudo, T, Tokiwa, H, Makishima, M, Yamada, S, Ikura, T, Ito, N. | | Deposit date: | 2012-05-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Combination of Triple Bond and Adamantane Ring on the Vitamin D Side Chain Produced Partial Agonists for Vitamin D Receptor.
J.Med.Chem., 2014
|
|
1UDY
 
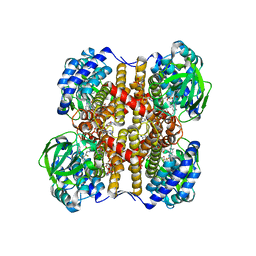 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
8WLS
 
 | |
5Y1F
 
 | | Monomeric L-threonine 3-dehydrogenase from metagenome database (NAD+ bound form) | | Descriptor: | NAD dependent epimerase/dehydratase family, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
3WMW
 
 | | GalE-like L-Threonine dehydrogenase from Cupriavidus necator (apo form) | | Descriptor: | CALCIUM ION, NAD dependent epimerase/dehydratase | | Authors: | Nakano, S, Okazaki, S, Tokiwa, H, Asano, Y. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of NAD+ and L-Threonine Induces Stepwise Structural and Flexibility Changes in Cupriavidus necator L-Threonine Dehydrogenase
J.Biol.Chem., 289, 2014
|
|
5Y1G
 
 | | Monomeric L-threonine 3-dehydrogenase from metagenome database (AKB and NADH bound form) | | Descriptor: | 2-AMINO-3-KETOBUTYRIC ACID, NAD dependent epimerase/dehydratase family, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
5Y1E
 
 | | monomeric L-threonine 3-dehydrogenase from metagenome database (L-Ser and NAD+ bound form) | | Descriptor: | NAD dependent epimerase/dehydratase family, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SERINE | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
5Y1D
 
 | | Monomeric L-threonine 3-dehydrogenase from metagenome database (apo form) | | Descriptor: | NAD dependent epimerase/dehydratase family | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
