1UL1
 
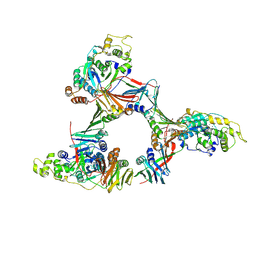 | | Crystal structure of the human FEN1-PCNA complex | | Descriptor: | Flap endonuclease-1, MAGNESIUM ION, Proliferating cell nuclear antigen | | Authors: | Sakurai, S, Kitano, K, Yamaguchi, H, Hamada, K, Okada, K, Fukuda, K, Uchida, M, Ohtsuka, E, Morioka, H, Hakoshima, T. | | Deposit date: | 2003-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for recruitment of human flap endonuclease 1 to PCNA
EMBO J., 24, 2005
|
|
6LFW
 
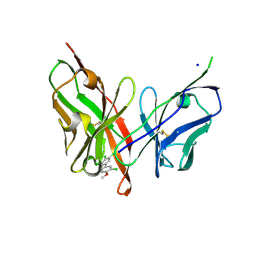 | |
6LCS
 
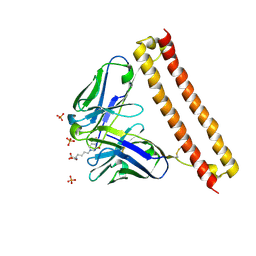 | | Crystal structure of 73MuL9 Fv-clasp fragment in complex with GA-pyridine analogue | | Descriptor: | (2~{S})-6-[4-(hydroxymethyl)-3-oxidanyl-pyridin-1-ium-1-yl]-2-(phenylmethoxycarbonylamino)hexanoic acid, PHOSPHATE ION, VH-SARAH, ... | | Authors: | Nakamura, T, Takagi, J, Yamagata, Y, Morioka, H. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of a single-chain Fv antibody specific for GA-pyridine, an advanced glycation end-product (AGE), elucidated using biophysical techniques and synthetic antigen analogues.
J.Biochem., 170, 2021
|
|
6LFV
 
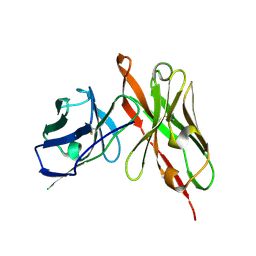 | |
1X1S
 
 | | Crystal structure of M-Ras in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras | | Authors: | Ye, M, Shima, F, Muraoka, S, Liao, J, Okamoto, H, Yamamoto, M, Tamura, A, Yagi, N, Ueki, T, Kataoka, T. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of M-Ras Reveals a GTP-bound "Off" State Conformation of Ras Family Small GTPases
J.Biol.Chem., 280, 2005
|
|
1X1R
 
 | | Crystal structure of M-Ras in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein M-Ras | | Authors: | Ye, M, Shima, F, Muraoka, S, Liao, J, Okamoto, H, Yamamoto, M, Tamura, A, Yagi, N, Ueki, T, Kataoka, T. | | Deposit date: | 2005-04-12 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of M-Ras Reveals a GTP-bound "Off" State Conformation of Ras Family Small GTPases
J.Biol.Chem., 280, 2005
|
|
2DKC
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the substrate complex | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2DKD
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the product complex | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-galactopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
1PIP
 
 | | CRYSTAL STRUCTURE OF PAPAIN-SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE COMPLEX AT 1.7 ANGSTROMS RESOLUTION: NONCOVALENT BINDING MODE OF A COMMON SEQUENCE OF ENDOGENOUS THIOL PROTEASE INHIBITORS | | Descriptor: | Papain, SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE | | Authors: | Yamamoto, A, Tomoo, K, Doi, M, Ohishi, H, Inoue, M, Ishida, T, Yamamoto, D, Tsuboi, S, Okamoto, H, Okada, Y. | | Deposit date: | 1992-10-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of papain-succinyl-Gln-Val-Val-Ala-Ala-p-nitroanilide complex at 1.7-A resolution: noncovalent binding mode of a common sequence of endogenous thiol protease inhibitors.
Biochemistry, 31, 1992
|
|
8GW8
 
 | | the human PTH1 receptor bound to an intracellular biased agonist | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Okamoto, H.H, Nureki, O. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Class B1 GPCR activation by an intracellular agonist.
Nature, 618, 2023
|
|
2DKA
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the apo-form | | Descriptor: | Phosphoacetylglucosamine mutase | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2OP3
 
 | | The structure of cathepsin S with a novel 2-arylphenoxyacetaldehyde inhibitor derived by the Substrate Activity Screening (SAS) method | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-[(2',3',4'-TRIFLUOROBIPHENYL-2-YL)OXY]ETHANOL, Cathepsin S, ... | | Authors: | Spraggon, G, Inagaki, H, Tsuruoka, H, Hornsby, M, Lesley, S.A, Ellman, J.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization and optimization of selective, nonpeptidic inhibitors of cathepsin S with an unprecedented binding mode.
J.Med.Chem., 50, 2007
|
|
5X3Z
 
 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
1NFT
 
 | | OVOTRANSFERRIN, N-TERMINAL LOBE, IRON LOADED OPEN FORM | | Descriptor: | FE (III) ION, NITRILOTRIACETIC ACID, PROTEIN (OVOTRANSFERRIN), ... | | Authors: | Mizutani, K, Yamashita, H, Kurokawa, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
1D8K
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1D8J
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1VD4
 
 | | Solution structure of the zinc finger domain of TFIIE alpha | | Descriptor: | Transcription initiation factor IIE, alpha subunit, ZINC ION | | Authors: | Okuda, M, Tanaka, A, Arai, Y, Satoh, M, Okamura, H, Nagadoi, A, Hanaoka, F, Ohkuma, Y, Nishimura, Y. | | Deposit date: | 2004-03-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc finger structure in the large subunit of human general transcription factor TFIIE.
J.Biol.Chem., 279, 2004
|
|
1J1R
 
 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Antiviral Protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1S
 
 | | Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Formycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral Protein S, FORMYCIN-5'-MONOPHOSPHATE | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1Q
 
 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1JOY
 
 | | SOLUTION STRUCTURE OF THE HOMODIMERIC DOMAIN OF ENVZ FROM ESCHERICHIA COLI BY MULTI-DIMENSIONAL NMR. | | Descriptor: | PROTEIN (ENVZ_ECOLI) | | Authors: | Tomomori, C, Tanaka, T, Dutta, R, Park, H, Saha, S.K, Zhu, Y, Ishima, R, Liu, D, Tong, K.I, Kurokawa, H, Qian, H, Inouye, M, Ikura, M. | | Deposit date: | 1998-12-28 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ.
Nat.Struct.Biol., 6, 1999
|
|
4BWQ
 
 | | Crystal structure of U5-15kD in a complex with PQBP1 | | Descriptor: | POLYGLUTAMINE-BINDING PROTEIN 1, THIOREDOXIN-LIKE PROTEIN 4A | | Authors: | Mizuguchi, M, Obita, T, Serita, T, Kojima, R, Morimoto, T, Nabeshima, Y, Okazawa, H. | | Deposit date: | 2013-07-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations in the Pqbp1 Gene Prevent its Interaction with the Spliceosomal Protein U5-15Kd.
Nat.Commun., 5, 2014
|
|
2SSP
 
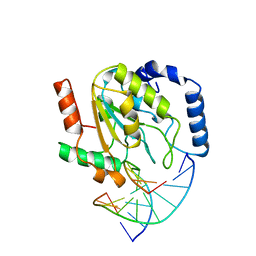 | | LEUCINE-272-ALANINE URACIL-DNA GLYCOSYLASE BOUND TO ABASIC SITE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(AAB)P*AP*TP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
2IUW
 
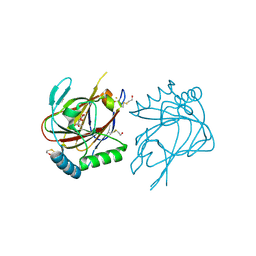 | | Crystal structure of human ABH3 in complex with iron ion and 2- oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ALKYLATED REPAIR PROTEIN ALKB HOMOLOG 3, BETA-MERCAPTOETHANOL, ... | | Authors: | Sundheim, O, Vagbo, C.B, Bjoras, M, deSousa, M.M.L, Talstad, V, Aas, P.A, Drablos, F, Krokan, H.E, Tainer, J.A, Slupphaug, G. | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human Abh3 Structure and Key Residues for Oxidative Demethylation to Reverse DNA/RNA Damage.
Embo J., 25, 2006
|
|
