1HSS
 
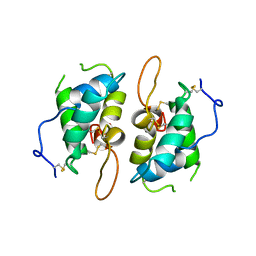 | | 0.19 ALPHA-AMYLASE INHIBITOR FROM WHEAT | | Descriptor: | 0.19 ALPHA-AMYLASE INHIBITOR | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tertiary and quaternary structures of 0.19 alpha-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 A resolution.
Biochemistry, 36, 1997
|
|
1WKV
 
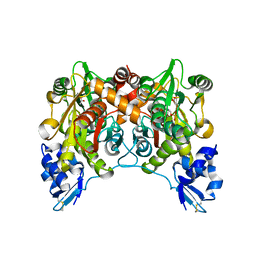 | | Crystal structure of O-phosphoserine sulfhydrylase | | Descriptor: | ACETATE ION, PYRIDOXAL-5'-PHOSPHATE, cysteine synthase | | Authors: | Oda, Y, Mino, K, Ishikawa, K, Ataka, M. | | Deposit date: | 2004-06-09 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional Structure of a New Enzyme, O-Phosphoserine Sulfhydrylase, involved in l-Cysteine Biosynthesis by a Hyperthermophilic Archaeon, Aeropyrum pernix K1, at 2.0A Resolution
J.Mol.Biol., 351, 2005
|
|
7ERV
 
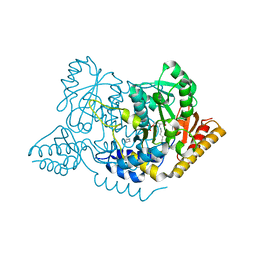 | | Crystal structure of L-histidine decarboxylase (C57S/C101V/C282V mutant) from Photobacterium phosphoreum | | Descriptor: | Histidine decarboxylase, IMIDAZOLE | | Authors: | Oda, Y, Nakata, K, Yamaguchi, H, Kashiwagi, T, Miyano, H, Mizukoshi, T. | | Deposit date: | 2021-05-07 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the enhanced thermostability of cysteine substitution mutants of L-histidine decarboxylase from Photobacterium phosphoreum.
J.Biochem., 171, 2022
|
|
7ERU
 
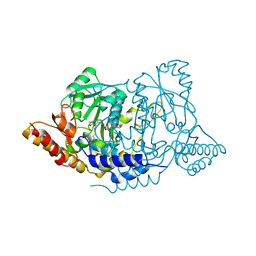 | | Crystal structure of L-histidine decarboxylase (C57S mutant) from Photobacterium phosphoreum | | Descriptor: | Histidine decarboxylase | | Authors: | Oda, Y, Nakata, K, Yamaguchi, H, Kashiwagi, T, Miyano, H, Mizukoshi, T. | | Deposit date: | 2021-05-07 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into the enhanced thermostability of cysteine substitution mutants of L-histidine decarboxylase from Photobacterium phosphoreum.
J.Biochem., 171, 2022
|
|
1C8N
 
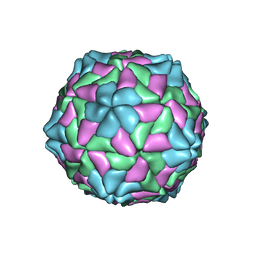 | | TOBACCO NECROSIS VIRUS | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 2000-05-20 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of tobacco necrosis virus at 2.25 A resolution.
J.Mol.Biol., 300, 2000
|
|
7YMJ
 
 | | Cryo-EM structure of alpha1AAR-Nb6 complex bound to tamsulosin | | Descriptor: | Nb6, Tamsulosin, alpha1A-adrenergic receptor | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
7YMH
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to noradrenaline | | Descriptor: | Nb29, Noradrenaline, alpha1A-adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
7YM8
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to oxymetazoline | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Oxymetazoline, alpha1A adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
5CL2
 
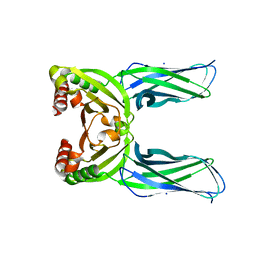 | | Crystal structure of Spo0M, sporulation control protein, from Bacillus subtilis. | | Descriptor: | SODIUM ION, Sporulation-control protein spo0M | | Authors: | Sonoda, Y, Mizutani, K, Mikami, B. | | Deposit date: | 2015-07-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Spo0M, a sporulation-control protein from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5BW7
 
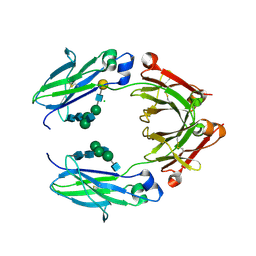 | | Crystal structure of nonfucosylated Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Isoda, Y, Yagi, H, Satoh, T, Shibata-Koyama, M, Masuda, K, Satoh, M, Kato, K, Iida, S. | | Deposit date: | 2015-06-06 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Importance of the Side Chain at Position 296 of Antibody Fc in Interactions with Fc gamma RIIIa and Other Fc gamma Receptors
Plos One, 10, 2015
|
|
1FZZ
 
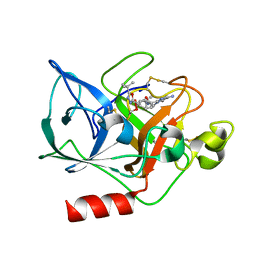 | | THE CRYSTAL STRUCTURE OF THE COMPLEX OF NON-PEPTIDIC INHIBITOR ONO-6818 AND PORCINE PANCREATIC ELASTASE. | | Descriptor: | 2-(5-AMINO-6-OXO-2-PHENYL-6H-PYRIMIDIN-1-YL)-N-[2-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)-1-(METHYLETHYL)-2-HYDROXYETHYL]ACETAMIDE, ELASTASE 1 | | Authors: | Odagaki, Y, Ohmoto, K, Matsuoka, S, Hamanaka, N, Nakai, H, Toda, M, Katsuya, Y. | | Deposit date: | 2000-10-04 | | Release date: | 2001-10-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The crystal structure of the complex of non-peptidic inhibitor of human neutrophil elastase ONO-6818 and porcine pancreatic elastase.
Bioorg.Med.Chem., 9, 2001
|
|
1AUG
 
 | | CRYSTAL STRUCTURE OF THE PYROGLUTAMYL PEPTIDASE I FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | PYROGLUTAMYL PEPTIDASE-1 | | Authors: | Odagaki, Y, Hayashi, A, Okada, K, Hirotsu, K, Kabashima, T, Ito, K, Yoshimoto, T, Tsuru, D, Sato, M, Clardy, J. | | Deposit date: | 1997-08-26 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pyroglutamyl peptidase I from Bacillus amyloliquefaciens reveals a new structure for a cysteine protease.
Structure Fold.Des., 7, 1999
|
|
1AUE
 
 | |
1BTP
 
 | | UNIQUE BINDING OF A NOVEL SYNTHETIC INHIBITOR, N-[3-[4-[4-(AMIDINOPHENOXY)-CARBONYL]PHENYL]-2-METHYL-2-PROPENOYL]-N-ALLYLGLYCINE METHANESULFONATE TO BOVINE TRYPSIN, REVEALED BY THE CRYSTAL STRUCTURE OF THE COMPLEX | | Descriptor: | BETA-TRYPSIN, CALCIUM ION | | Authors: | Odagaki, Y, Nakai, H, Senokuchi, K, Kawamura, M, Hamanaka, N, Nakamura, M, Tomoo, K, Ishida, T. | | Deposit date: | 1995-08-11 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique binding of a novel synthetic inhibitor, N-[3-[4-[4-(amidinophenoxy)carbonyl]phenyl]-2-methyl-2-propenoyl]- N-allylglycine methanesulfonate, to bovine trypsin, revealed by the crystal structure of the complex.
Biochemistry, 34, 1995
|
|
7WCE
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with (2S)-2-(tert-Butoxy)-2-(10-fluoro-2-(2-hydroxy-4-methylphenyl)-1,4-dimethyl-5-(methylsulfonyl)-5,6-dihydrophenanthridin-3-yl)acetic acid | | Descriptor: | (2S)-2-[10-fluoranyl-1,4-dimethyl-2-(4-methyl-2-oxidanyl-phenyl)-5-methylsulfonyl-6H-phenanthridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, GLYCEROL, Integrase catalytic, ... | | Authors: | Taoda, Y, Sekiguchi, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of tricyclic HIV-1 integrase-LEDGF/p75 allosteric inhibitors by intramolecular direct arylation reaction.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
5YFI
 
 | | Crystal structure of the anti-human prostaglandin E receptor EP4 antibody Fab fragment | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, ZINC ION | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YHL
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and an antagonist Br-derivative | | Descriptor: | 4-[2-[[(2R)-2-(4-bromanylnaphthalen-1-yl)propanoyl]amino]-4-cyano-phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-28 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YWY
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 | | Descriptor: | 4-[4-cyano-2-[[(2R)-2-(4-fluoranylnaphthalen-1-yl)propanoyl]amino]phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
2RPS
 
 | | Solution structure of a novel insect chemokine isolated from integument | | Descriptor: | Chemokine | | Authors: | Kamiya, M, Nakatogawa, S, Oda, Y, Kamijima, T, Aizawa, T, Demura, M, Hayakawa, Y, Kawano, K. | | Deposit date: | 2008-07-28 | | Release date: | 2009-06-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A novel peptide mediates aggregation and migration of hemocytes from an insect
Curr.Biol., 19, 2009
|
|
8H9X
 
 | |
8HA2
 
 | |
8H9Y
 
 | |
8HA1
 
 | |
8XZ2
 
 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
4X42
 
 | | Crystal structure of DEN4 ED3 mutant with epitope two residues substituted from DEN3 ED3 | | Descriptor: | Envelope protein E, SULFATE ION | | Authors: | Kulkarni, M.R, Islam, M.M, Numoto, N, Elahi, M.M, Ito, N, Kuroda, Y. | | Deposit date: | 2014-12-02 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and biophysical analysis of sero-specific immune responses using epitope grafted Dengue ED3 mutants.
Biochim.Biophys.Acta, 1854, 2015
|
|
