8XBG
 
 | |
8WUT
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUS
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUV
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUU
 
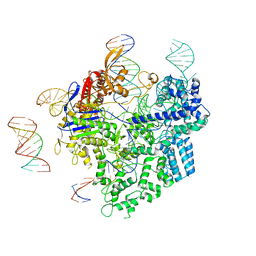 | | SpCas9-pegRNA-target DNA complex (pre-initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (34-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
7YTB
 
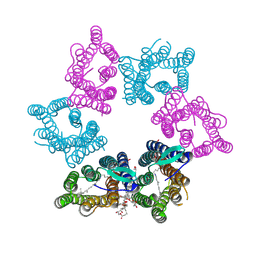 | | Crystal structure of Kin4B8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Kin4B8, RETINAL | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2022-08-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
1QQT
 
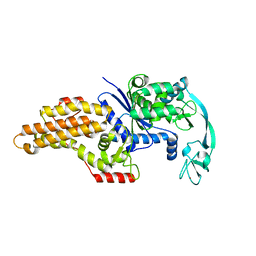 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Mechulam, Y, Schmitt, E, Maveyraud, L, Zelwer, C, Nureki, O, Yokoyama, S, Konno, M, Blanquet, S. | | Deposit date: | 1999-06-08 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features.
J.Mol.Biol., 294, 1999
|
|
6LMU
 
 | | Cryo-EM structure of the human CALHM2 | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6M04
 
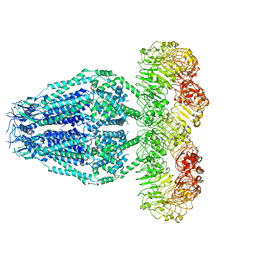 | | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8D | | Authors: | Nakamura, R, Kasuya, G, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol, 3, 2020
|
|
6LMX
 
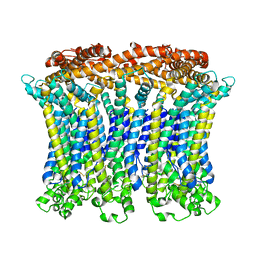 | | Cryo-EM structure of the CALHM chimeric construct (9-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LRY
 
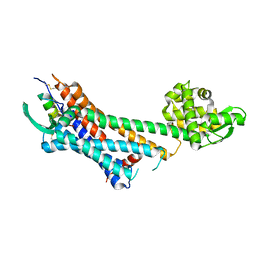 | | Crystal structure of human endothelin ETB receptor in complex with sarafotoxin S6b | | Descriptor: | Endothelin receptor type B,Endolysin,Endothelin receptor type B, Sarafotoxin-B | | Authors: | Izume, T, Miyauchi, H, Shihoya, W, Nureki, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human endothelin ETBreceptor in complex with sarafotoxin S6b.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
6LMV
 
 | | Cryo-EM structure of the C. elegans CLHM-1 | | Descriptor: | Calcium homeostasis modulator protein | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMW
 
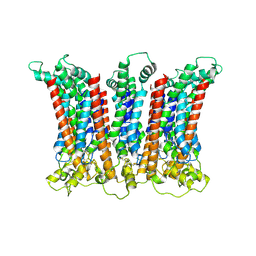 | | Cryo-EM structure of the CALHM chimeric construct (8-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
7BTW
 
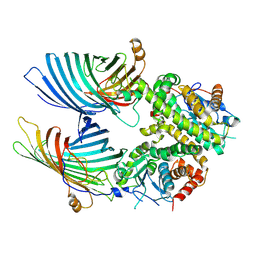 | | The mitochondrial SAM complex from S.cere | | Descriptor: | Mitochondrial outer membrane beta-barrel protein, SAM37 isoform 1, Sorting assembly machinery 35 kDa subunit | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTX
 
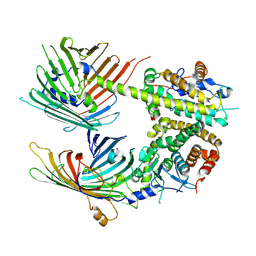 | | The mitochondrial SAM-Mdm10 supercomplex in GDN micelle from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTY
 
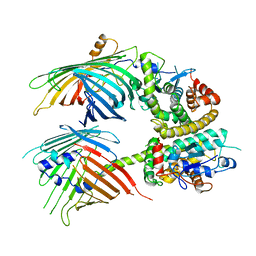 | | The mitochondrial SAM-Mdm10 supercomplex in Nanodisc from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7YU5
 
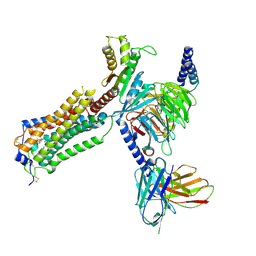 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU6
 
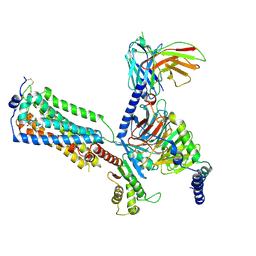 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state2 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU3
 
 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU8
 
 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU7
 
 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state3 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
1WOY
 
 | |
1WY5
 
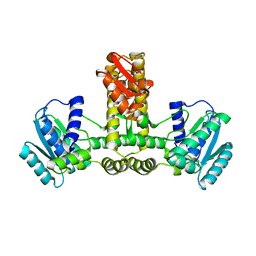 | | Crystal structure of isoluecyl-tRNA lysidine synthetase | | Descriptor: | Hypothetical UPF0072 protein AQ_1887 | | Authors: | Nakanishi, K, Fukai, S, Ikeuchi, Y, Soma, A, Sekine, Y, Suzuki, T, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for lysidine formation by ATP pyrophosphatase accompanied by a lysine-specific loop and a tRNA-recognition domain.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4PDN
 
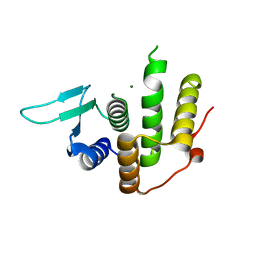 | | Crystal structure of E. coli YfcM | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation.
Nucleic Acids Res., 42, 2014
|
|
4X5M
 
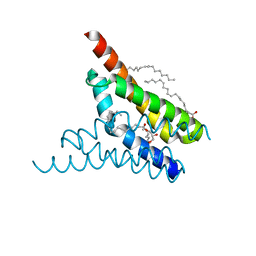 | | Crystal structure of SemiSWEET in the inward-open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Lee, Y, Nishizawa, T, Yamashita, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter
Nat Commun, 6, 2015
|
|
