3ETZ
 
 | | Crystal structure of bacterial adhesin FadA L76A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
8F6G
 
 | |
7SHH
 
 | |
2GL2
 
 | | Crystal structure of the tetra mutant (T66G,R67G,F68G,Y69G) of bacterial adhesin FadA | | Descriptor: | adhesion A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2006-04-04 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary X-ray data of the FadA adhesin from Fusobacterium nucleatum.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
6B5D
 
 | |
6B5C
 
 | |
7S5U
 
 | |
5CYA
 
 | | Crystal structure of Arl2 GTPase-activating protein tubulin cofactor C (TBCC) | | Descriptor: | SULFATE ION, Tubulin-specific chaperone C | | Authors: | Nithianantham, S, Le, S, Seto, E, Jia, W, Leary, J, Corbett, K.D, Moore, J.K, Al-Bassam, J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble alpha beta-tubulin pool for microtubule dynamics.
Elife, 4, 2015
|
|
7M2E
 
 | | Crystal structure of BPTF bromodomain in complex with CB02-092 | | Descriptor: | 4-chloro-5-{4-[2-(dimethylamino)ethyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Nithianantham, S, Fischer, M. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition
J.Med.Chem., 64, 2021
|
|
3ETY
 
 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETX
 
 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETW
 
 | | Crystal Structure of bacterial adhesin FadA | | Descriptor: | Adhesin A, THIOCYANATE ION | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
6MZG
 
 | |
6MZE
 
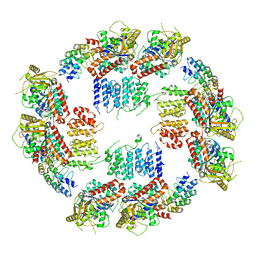 | |
6MZF
 
 | |
3BXZ
 
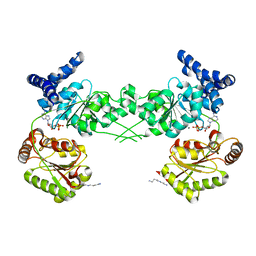 | | Crystal structure of the isolated DEAD motor domains from Escherichia coli SecA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Preprotein translocase subunit secA, ... | | Authors: | Nithianantham, S, Namjoshi, S, Shilton, B.H. | | Deposit date: | 2008-01-15 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Analysis of the isolated SecA DEAD motor suggests a mechanism for chemical-mechanical coupling.
J.Mol.Biol., 383, 2008
|
|
2B5U
 
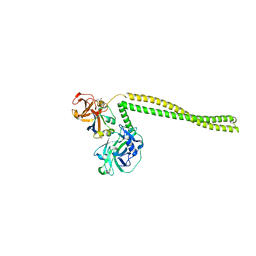 | | Crystal Structure Of Colicin E3 V206C Mutant In Complex With Its Immunity Protein | | Descriptor: | CITRIC ACID, Colicin E3, Colicin E3 immunity protein | | Authors: | Nallini Vijayarangan, A, Nithianantham, S, Nan, W, Jakes, K, Shoham, M. | | Deposit date: | 2005-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Of Colicin E3 In Complex With Its Immunity Protein
TO BE PUBLISHED
|
|
4PXT
 
 | |
7ULK
 
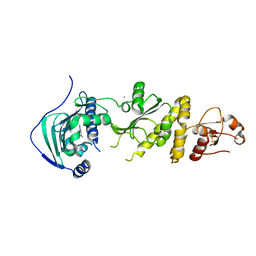 | | Human TRAP1 NM in complex with 42C | | Descriptor: | CALCIUM ION, Heat shock protein 75 kDa, mitochondrial, ... | | Authors: | Stachowski, T.R, Nithianantham, S, Vanarotti, M, Fischer, M. | | Deposit date: | 2022-04-05 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Pan-HSP90 ligand binding reveals isoform-specific differences in plasticity and water networks.
Protein Sci., 32, 2023
|
|
4PXU
 
 | | Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bipolar kinesin KRP-130 | | Authors: | Scholey, J.E, Nithianantham, S, Scholey, J.M, Al-Bassam, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers.
Elife, 3, 2014
|
|
7ULJ
 
 | | Hsp90b N-terminal domain in complex with 42C | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Heat shock protein HSP 90-beta, ... | | Authors: | Stachowski, T.R, Nithianantham, S, Vanarotti, M, Fischer, M. | | Deposit date: | 2022-04-05 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Pan-HSP90 ligand binding reveals isoform-specific differences in plasticity and water networks.
Protein Sci., 32, 2023
|
|
7ULL
 
 | | Human Grp94 N-terminal domain in complex with 42C | | Descriptor: | DIMETHYL SULFOXIDE, Endoplasmin, GLYCEROL, ... | | Authors: | Stachowski, T.R, Nithianantham, S, Vanarotti, M, Fischer, M. | | Deposit date: | 2022-04-05 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Pan-HSP90 ligand binding reveals isoform-specific differences in plasticity and water networks.
Protein Sci., 32, 2023
|
|
6VPP
 
 | | Cryo-EM structure of microtubule-bound KLP61F motor with tail domain in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp61F, ... | | Authors: | Bodrug, T, Wilson-Kubalek, E.M, Nithianantham, S, Debs, G, Sindelar, C.V, Milligan, R, Al-Bassam, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding.
Elife, 9, 2020
|
|
6VPO
 
 | | Cryo-EM structure of microtubule-bound KLP61F motor domain in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp61F, ... | | Authors: | Bodrug, T, Wilson-Kubalek, E.M, Nithianantham, S, Debs, G, Sindelar, C.V, Milligan, R, Al-Bassam, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding.
Elife, 9, 2020
|
|
6WTQ
 
 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-4-{[4-(1-propyl-1H-pyrazol-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}benzamide, ... | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79968476 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
