7A8P
 
 | | Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor. | | Descriptor: | (3~{R})-1-[(2~{R})-2-[4-(2-chloranyl-4-fluoranyl-phenyl)-2-oxidanylidene-chromen-7-yl]oxypropanoyl]piperidine-3-carboxylic acid, DNA-directed RNA polymerase, mitochondrial | | Authors: | Hillen, H.S, Bonekamp, N, Peter, B, Felser, A, Bergbrede, T, Choidas, A, Horn, M, Unger, A, di Lucrezia, R, Atanassov, I, Li, X, Koch, U, Menninger, S, Boros, J, Habenberger, P, Giavalisco, P, Cramer, P, Denzel, M, Nussbaumer, P, Klebl, B, Falkenberg, M, Gustafsson, C.M, Larsson, N.G. | | Deposit date: | 2020-08-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Small-molecule inhibitors of human mitochondrial DNA transcription.
Nature, 588, 2020
|
|
7N9O
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Aliphatic SERD S-C10(15) | | Descriptor: | 10-{[3,17beta-dihydroxyestra-1,3,5(10)-trien-7beta-yl]sulfanyl}-N-methyl-N-propyldecanamide, Estrogen receptor | | Authors: | Diennet, M, El Ezzy, M, Thiombane, K, Cotnoir-White, D, Poupart, J, Gao, Z, Mendoza, S.R, Marinier, A, Gleason, J, Mader, S.C, Fanning, S.W. | | Deposit date: | 2021-06-18 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Aliphatic SERD S-C10(15)
To Be Published
|
|
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-08 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
8OV5
 
 | | PERIDININ-CHLOROPHYLL-PROTEIN OF AMPHIDINIUM CARTERAE, 100K | | Descriptor: | CHLOROPHYLL A, PERIDININ, Peridinin-chlorophyll a-binding protein 1, ... | | Authors: | Hofmann, E, Johanning, S. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and spectroscopic characterization of the peridinin-chlorophyll a-protein (PCP) complex from Heterocapsa pygmaea (HPPCP).
Biochim Biophys Acta Bioenerg, 1866, 2024
|
|
7SFO
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with 3-(((2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)amino)methyl)phenol and GRIP Peptide | | Descriptor: | 3-{[(2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)amino]methyl}phenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Joyner, C, Reddy, V.K, Wilson, T, Fanning, S.W. | | Deposit date: | 2021-10-04 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estrogen Receptor Alpha
To Be Published
|
|
7RKE
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with 4-(((2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)amino)methyl)phenol and GRIP Peptide | | Descriptor: | 4-{[(2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)amino]methyl}phenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Willson, T.M, Fanning, S.W. | | Deposit date: | 2021-07-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A New Chemotype of Chemically Tractable Nonsteroidal Estrogens Based on a Thieno[2,3- d ]pyrimidine Core.
Acs Med.Chem.Lett., 13, 2022
|
|
7R62
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with a Desmethyl ICI164,384 Derivative | | Descriptor: | 11-[3,17beta-dihydroxyestra-1,3,5(10)-trien-7beta-yl]-N-methyl-N-propylundecanamide, Estrogen receptor | | Authors: | Diennet, M, El Ezzy, M, Thiombane, K, Cotnoir-White, D, Poupart, J, Gao, Z, Mendoza Sanchez, R, Marinier, A, Gleason, J, Greene, G.L, Mader, S.C, Fanning, S.W. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain in Complex with a Desmethyl ICI164,384 Derivative
To Be Published
|
|
7RNM
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with 2-(2-Chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)isoindolin-5-ol and GRIP Peptide | | Descriptor: | 2-(2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-isoindol-5-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Willson, T.M, Fanning, S.W. | | Deposit date: | 2021-07-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of the Binding and Activation of Estrogen Receptor alpha by Phenolic Thieno[2,3-d]pyrimidines
Helv.Chim.Acta, 106, 2023
|
|
7TE7
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with RAD1901 | | Descriptor: | (6R)-6-{2-[ethyl({4-[2-(ethylamino)ethyl]phenyl}methyl)amino]-4-methoxyphenyl}-5,6,7,8-tetrahydronaphthalen-2-ol, Estrogen receptor | | Authors: | Joiner, C, Hancock, G, Young, K, Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
5DX3
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Stapled Peptide SRC2-P3 and Estradiol | | Descriptor: | CHLORIDE ION, ESTRADIOL, Estrogen receptor, ... | | Authors: | Speltz, T.E, Fanning, S.W, Mayne, C.G, Tajkhorshid, E, Greene, G.L, Moore, T.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.0903 Å) | | Cite: | Stapled Peptides with gamma-Methylated Hydrocarbon Chains for the Estrogen Receptor/Coactivator Interaction.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3QSK
 
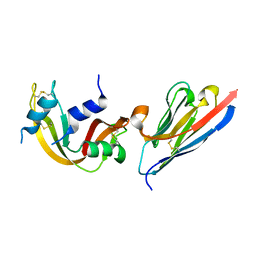 | | 5 Histidine Variant of the anti-RNase A VHH in Complex with RNAse A | | Descriptor: | Engineered 5 Histidine anti-RNase A Camelid VHH Antibody Domain Variant, Ribonuclease pancreatic | | Authors: | Murtaugh, M.L, Fanning, S.W, Sharma, T.M, Terry, A.M, Horn, J.R. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-10 | | Last modified: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A combinatorial histidine scanning library approach to engineer highly pH-dependent protein switches.
Protein Sci., 20, 2011
|
|
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
4ATJ
 
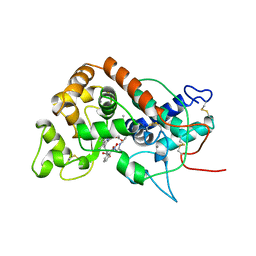 | | DISTAL HEME POCKET MUTANT (H42E) OF RECOMBINANT HORSERADISH PEROXIDASE IN COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PROTEIN (PEROXIDASE C1A), ... | | Authors: | Meno, K, Jennings, S, Smith, A.T, Henriksen, A, Gajhede, M. | | Deposit date: | 1999-04-19 | | Release date: | 2002-10-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the two horseradish peroxidase catalytic residue variants H42E and R38S/H42E: implications for the catalytic cycle.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7T2X
 
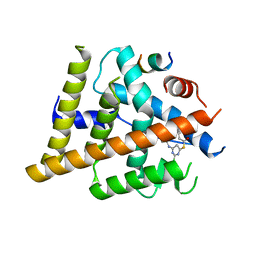 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with 2-chloro-4-((4-hydroxybenzyl)amino)-5-phenylthieno[2,3-d]pyrimidin-6-ol and GRIP Peptide | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, S-(2-chloro-6-{[(4-hydroxyphenyl)methyl]amino}pyrimidin-4-yl) phenylethanethioate | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Wilson, T.M, Fanning, S.W. | | Deposit date: | 2021-12-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A New Chemotype of Chemically Tractable Nonsteroidal Estrogens Based on a Thieno[2,3- d ]pyrimidine Core.
Acs Med.Chem.Lett., 13, 2022
|
|
7RCU
 
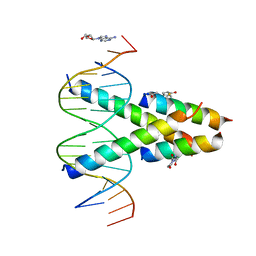 | | Synthetic Max homodimer mimic in complex with DNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2-(2,5-dioxopyrrolidin-1-yl)acetamide, ACETAMIDE, ... | | Authors: | Speltz, T, Qiao, Z, Shangguan, S, Fanning, S, Greene, J, Moellering, R. | | Deposit date: | 2021-07-08 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains.
Nat.Biotechnol., 41, 2023
|
|
1AIH
 
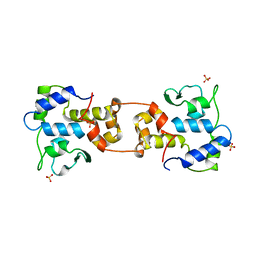 | | CATALYTIC DOMAIN OF BACTERIOPHAGE HP1 INTEGRASE | | Descriptor: | HP1 INTEGRASE, MAGNESIUM ION, SULFATE ION | | Authors: | Hickman, A.B, Waninger, S, Scocca, J.J, Dyda, F. | | Deposit date: | 1997-04-17 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution.
Cell(Cambridge,Mass.), 89, 1997
|
|
5JAF
 
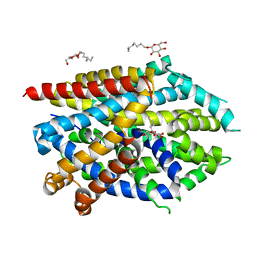 | | LeuT Na+-free Return State, C2 form at pH 5 | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.021 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
5JAG
 
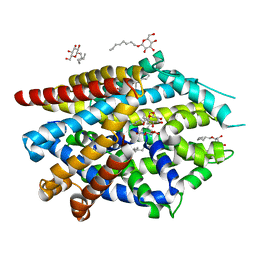 | | LeuT T354H mutant in the outward-oriented, Na+-free Return State | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
4AXP
 
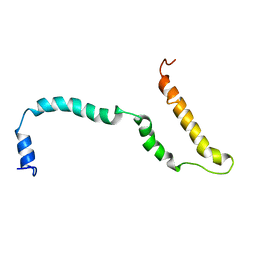 | | NMR structure of Hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast. | | Descriptor: | 12 KDA HEAT SHOCK PROTEIN | | Authors: | Herbert, A.P, Riesen, M, Bloxam, L, Kosmidou, E, Wareing, B.M, Johnson, J.R, Phelan, M.M, Pennington, S.R, Lian, L.Y, Morgan, A. | | Deposit date: | 2012-06-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Hsp12, a Protein Induced by and Required for Dietary Restriction-Induced Lifespan Extension in Yeast.
Plos One, 7, 2012
|
|
7UJC
 
 | |
7UJ8
 
 | |
7UJF
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with a Methylated Lasofoxifene Derivative with Selective Estrogen Receptor Degrader Properties | | Descriptor: | (5R,6S)-5-(4-{2-[(3S)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-6-phenyl-5,6,7,8-tetrahydronaphthalen-2-ol, Estrogen receptor | | Authors: | Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-03-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells.
Elife, 11, 2022
|
|
7UJW
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with a Methylated Lasofoxifene Derivative that Possesses Selective Estrogen Receptor Degrader Activities | | Descriptor: | (5R,6S)-5-(4-{2-[(3S)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-6-phenyl-5,6,7,8-tetrahydronaphthalen-2-ol, Estrogen receptor | | Authors: | Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-03-31 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells.
Elife, 11, 2022
|
|
