8XWX
 
 | |
6QWU
 
 | | 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus at pH 5.5 with Mn2+ and CoA. | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-03-06 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
6QXQ
 
 | |
6QXR
 
 | | 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus at pH 8.5 with Mn2+ and CoA. | | Descriptor: | CALCIUM ION, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
6RCX
 
 | | Mycobacterial 4'-phosphopantetheinyl transferase PptAb in complex with the ACP domain of PpsC. | | Descriptor: | CACODYLATE ION, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
6QYG
 
 | |
6QYF
 
 | |
8U77
 
 | | Crystal structure of Taf14 in complex with Yng1 | | Descriptor: | Protein YNG1, Transcription initiation factor TFIID subunit 14 | | Authors: | Nguyen, M.C, Wei, P.C, Zhang, G.Y, Kutateladze, T.G. | | Deposit date: | 2023-09-14 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular insight into interactions between the Taf14, Yng1 and Sas3 subunits of the NuA3 complex.
Nat Commun, 15, 2024
|
|
1EQ9
 
 | | CRYSTAL STRUCTURE OF FIRE ANT CHYMOTRYPSIN COMPLEXED TO PMSF | | Descriptor: | CHYMOTRYPSIN, phenylmethanesulfonic acid | | Authors: | Botos, I, Meyer, E, Nguyen, M.H, Swanson, S.M, Meyer, E.F. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of an insect chymotrypsin.
J.Mol.Biol., 298, 2000
|
|
7UNN
 
 | | Thiol-disulfide oxidoreductase TsdA from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thioredoxin domain-containing protein | | Authors: | Osipiuk, J, Reardon-Robinson, M, Nguyen, M.T, Sanchez, B, Ton-That, H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-11 | | Release date: | 2022-04-20 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A cryptic oxidoreductase safeguards oxidative protein folding in Corynebacterium diphtheriae.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UNO
 
 | | Thiol-disulfide oxidoreductase TsdA, C129S mutant, from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Reardon-Robinson, M, Nguyen, M.T, Sanchez, B, Ton-That, H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-11 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thiol-disulfide oxidoreductase in Corynebacterium diphtheriae
To Be Published
|
|
8PII
 
 | | VHH Z70 mutant 3 in interaction with PHF6 Tau peptide | | Descriptor: | Microtubule-associated protein tau, VHH Z70 Mutant 3 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
4XRT
 
 | | Crystal structure of the di-domain ARO/CYC StfQ from the steffimycin biosynthetic pathway | | Descriptor: | FORMIC ACID, StfQ Aromatase/Cyclase | | Authors: | Tsai, S.C, Caldara-Festin, G.M, Jackson, D.R, Aguilar, S, Patel, A, Nguyen, M, Sasaki, E, Valentic, T.R, Barajas, J.F, Vo, M, Khanna, A, Liu, H.-W. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural and functional analysis of two di-domain aromatase/cyclases from type II polyketide synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XRW
 
 | | Crystal structure of the di-domain ARO/CYC BexL from the BE-7585A biosynthetic pathway | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BexL | | Authors: | Tsai, S.C, Caldara-Festin, G.M, Jackson, D.R, Aguilar, S, Patel, A, Nguyen, M, Sasaki, E, Valentic, T.R, Barajas, J.F, Vo, M, Khanna, A, Liu, H.-W. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional analysis of two di-domain aromatase/cyclases from type II polyketide synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8OPI
 
 | | VHH Z70 mutant 1 in interaction with PHF6 Tau peptide | | Descriptor: | PHF6 Tau peptide, VHH Z70 mutant 1 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-04-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
8OP0
 
 | | VHH Z70 mutant 20 in interaction with PHF6 Tau peptide | | Descriptor: | Tau PHF6 peptide, VHH Z70 mutant 20 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
4WV3
 
 | | Crystal structure of the anthranilate CoA ligase AuaEII in complex with anthranoyl-AMP | | Descriptor: | 5'-O-[(S)-[(2-aminobenzoyl)oxy](hydroxy)phosphoryl]adenosine, Anthranilate-CoA ligase | | Authors: | Tsai, S.C, Muller, R, Jackson, D.R, Tu, S.S, Nguyen, M, Barajas, J.F, Pistorious, D, Krug, D. | | Deposit date: | 2014-11-04 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structural Insights into Anthranilate Priming during Type II Polyketide Biosynthesis.
Acs Chem.Biol., 11, 2016
|
|
5MTF
 
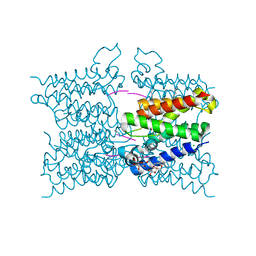 | | A modular route to novel potent and selective inhibitors of rhomboid intramembrane proteases | | Descriptor: | CHLORIDE ION, Rhomboid protease GlpG, inhibitor, ... | | Authors: | Ticha, A, Stanchev, S, Vinothkumar, K.R, Mikles, D.C, Pachl, P, Svehlova, K, Nguyen, M.T.N, Verhelst, S.H.L, Johnson, D, Bachovchin, D, Lepsik, M, Majer, P, Strisovsky, K. | | Deposit date: | 2017-01-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | General and Modular Strategy for Designing Potent, Selective, and Pharmacologically Compliant Inhibitors of Rhomboid Proteases.
Cell Chem Biol, 24, 2017
|
|
3HEI
 
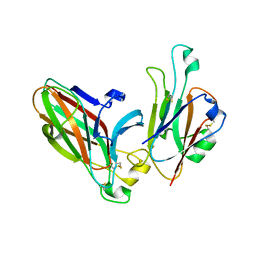 | | Ligand Recognition by A-Class Eph Receptors: Crystal Structures of the EphA2 Ligand-Binding Domain and the EphA2/ephrin-A1 Complex | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-05-08 | | Release date: | 2009-06-30 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
3HPN
 
 | | Ligand recognition by A-class EPH receptors: crystal structures of the EPHA2 ligand-binding domain and the EPHA2/EPHRIN-A1 complex | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-06-04 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
7BDW
 
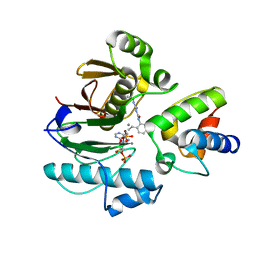 | | Crystal structure of mycobacterial PptAb in complex with ACP and compound 8918 | | Descriptor: | COENZYME A, MANGANESE (II) ION, N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea, ... | | Authors: | Carivenc, C, Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2020-12-22 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
1N2Y
 
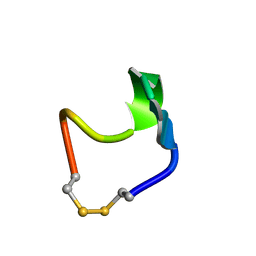 | | SOLUTION STRUCTURE OF SS-CYCLIZED CATESTATIN FRAGMENT FROM CHROMOGRANIN A | | Descriptor: | CATESTATIN | | Authors: | Preece, N.E, Nguyen, M, Mahata, M, Mahata, S.K, Mahapatra, N.R, Tsigelny, I, O'Connor, D.T. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences and activities of peptides from the catecholamine
release-inhibitory (catestatin) region of chromogranin A
Regul.Pept., 118, 2004
|
|
1Z8V
 
 | | The Structure of d(GGCCAATTGG) Complexed with Netropsin | | Descriptor: | (5'-D(*GP*GP*CP*CP*AP*AP*TP*TP*GP*G)-3'), NETROPSIN | | Authors: | Van Hecke, K, Nam, P.C, Nguyen, M.T, Van Meervelt, L. | | Deposit date: | 2005-03-31 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Netropsin interactions in the minor groove of d(GGCCAATTGG) studied by a combination of resolution enhancement and ab initio calculations.
Febs J., 272, 2005
|
|
