7LAF
 
 | | 15-lipoxygenase-2 loop mutant bound to imidazole-based inhibitor | | Descriptor: | 3-{[(4-methylphenyl)methyl]sulfanyl}-1-phenyl-1H-1,2,4-triazole, MANGANESE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX15B | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2021-01-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Kinetic and structural investigations of novel inhibitors of human epithelial 15-lipoxygenase-2.
Bioorg.Med.Chem., 46, 2021
|
|
3O8Y
 
 | | Stable-5-Lipoxygenase | | Descriptor: | Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Newcomer, M.E, Gilbert, N.C, Bartlett, S.G, Waight, M.T, Neau, D.B, Boeglin, W.E, Brash, A.R. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | The structure of human 5-lipoxygenase.
Science, 331, 2011
|
|
1EPA
 
 | |
1EPB
 
 | |
1BI9
 
 | | RETINAL DEHYDROGENASE TYPE TWO WITH NAD BOUND | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, RETINAL DEHYDROGENASE TYPE II | | Authors: | Newcomer, M.E, Lamb, A.L. | | Deposit date: | 1998-06-23 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity.
Biochemistry, 38, 1999
|
|
6N2W
 
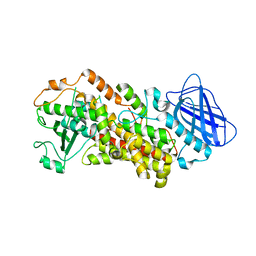 | | The structure of Stable-5-Lipoxygenase bound to NDGA | | Descriptor: | 4-[(2R,3S)-3-[(3,4-DIHYDROXYPHENYL)METHYL]-2-METHYLBUTYL]BENZENE-1,2-DIOL, Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2018-11-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products.
Nat.Chem.Biol., 16, 2020
|
|
6NCF
 
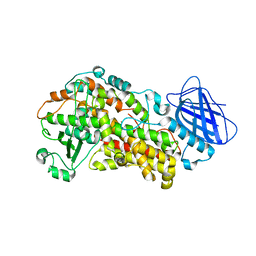 | | The structure of Stable-5-Lipoxygenase bound to AKBA | | Descriptor: | (3alpha,8alpha,17alpha,18alpha)-3-(acetyloxy)-11-oxours-12-en-23-oic acid, Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2018-12-11 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.871 Å) | | Cite: | Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products.
Nat.Chem.Biol., 16, 2020
|
|
1RBP
 
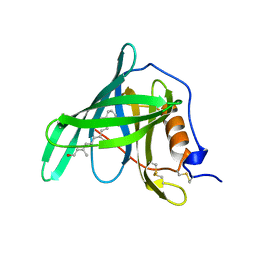 | |
4QWT
 
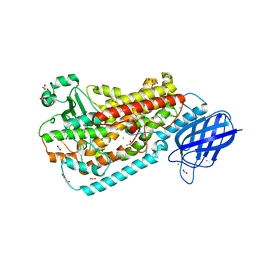 | |
5DD8
 
 | | The Crystal structure of HucR mutant (HucR-E48Q) from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, Transcriptional regulator, MarR family | | Authors: | Deochand, D.K, Perera, I.C, Crochet, R.B, Gilbert, N.C, Newcomer, M.E, Grove, A. | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Histidine switch controlling pH-dependent protein folding and DNA binding in a transcription factor at the core of synthetic network devices.
Mol Biosyst, 12, 2016
|
|
6AUM
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with trans-4-[4-(3-trifluoromethoxyphenyl-l-ureido)-cyclohexyloxy]-benzoic acid. | | Descriptor: | 4-{[trans-4-({[4-(trifluoromethoxy)phenyl]carbamoyl}amino)cyclohexyl]oxy}benzoic acid, Bifunctional epoxide hydrolase 2, CHLORIDE ION, ... | | Authors: | Kodani, S.D, Bahkta, S, Hwang, S.H, Pakhomova, S, Newcomer, M.E, Morisseau, C, Hammock, B. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification and optimization of soluble epoxide hydrolase inhibitors with dual potency towards fatty acid amide hydrolase.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
4J03
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with fulvestrant | | Descriptor: | (7beta,9beta,13alpha,17beta)-7-{9-[(R)-(4,4,5,5,5-pentafluoropentyl)sulfinyl]nonyl}estra-1(10),2,4-triene-3,17-diol, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Morisseau, C, Pakhomova, S, Hwang, S.H, Newcomer, M.E, Hammock, B.D. | | Deposit date: | 2013-01-30 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Inhibition of soluble epoxide hydrolase by fulvestrant and sulfoxides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KUM
 
 | | Structure of LSD1-CoREST-Tetrahydrofolate complex | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Calcutt, M.W, Newcomer, M.E, Wagner, C. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the histone lysine specific demethylase LSD1 complexed with tetrahydrofolate.
Protein Sci., 23, 2014
|
|
7TTJ
 
 | | Stable-5-LOX elongated Ha2 | | Descriptor: | Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Gilbert, N.C, Newcomer, M.E. | | Deposit date: | 2022-02-01 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Helical remodeling augments 5-lipoxygenase activity in the synthesis of proinflammatory mediators.
J.Biol.Chem., 298, 2022
|
|
7TTL
 
 | | Stable-5-LOX elongated Ha2 (4 copies ASU) | | Descriptor: | Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Gilbert, N.C, Newcomer, M.E. | | Deposit date: | 2022-02-01 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Helical remodeling augments 5-lipoxygenase activity in the synthesis of proinflammatory mediators.
J.Biol.Chem., 298, 2022
|
|
5UYT
 
 | | Crystal structure of ice binding protein from an Antarctic bacterium Flavobacteriaceae | | Descriptor: | Ice-binding protein, NITRATE ION | | Authors: | Wang, C, Pakhomova, S, Newcomer, M.E, Christner, B.C, Luo, B.-H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of antifreeze activity of a bacterial multi-domain antifreeze protein.
PLoS ONE, 12, 2017
|
|
4HAI
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with N-cycloheptyl-1-(mesitylsulfonyl)piperidine-4-carboxamide. | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, N-cycloheptyl-1-[(2,4,6-trimethylphenyl)sulfonyl]piperidine-4-carboxamide, ... | | Authors: | Pecic, S, Pakhomova, S, Newcomer, M.E, Morisseau, C, Hammock, B.D, Zhu, Z, Deng, S. | | Deposit date: | 2012-09-26 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis and structure-activity relationship of piperidine-derived non-urea soluble epoxide hydrolase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2AZT
 
 | | Crystal structure of H176N mutant of human Glycine N-Methyltransferase | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Luka, Z, Pakhomova, S, Luka, Y, Newcomer, M.E, Wagner, C. | | Deposit date: | 2005-09-12 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Destabilization of human glycine N-methyltransferase by H176N mutation.
Protein Sci., 16, 2007
|
|
2BAY
 
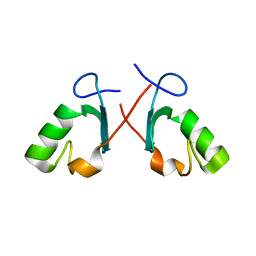 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
1X8L
 
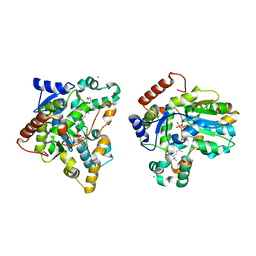 | | Crystal structure of retinol dehydratase in complex with all-trans-4-oxoretinol and inactive cofactor PAP | | Descriptor: | 4-OXORETINOL, ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
1X8K
 
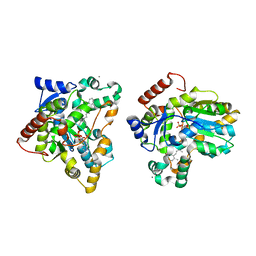 | | Crystal structure of retinol dehydratase in complex with anhydroretinol and inactive cofactor PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ANHYDRORETINOL, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
1X8J
 
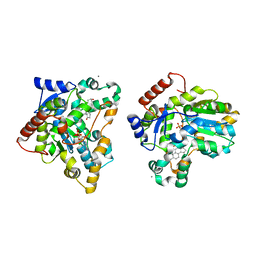 | | Crystal structure of retinol dehydratase in complex with androsterone and inactive cofactor PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Androsterone, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
2FBK
 
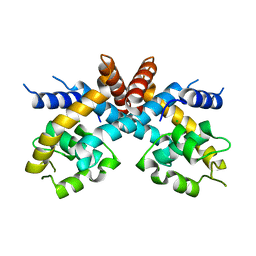 | | The Crystal Structure of HucR from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, transcriptional regulator, MarR family | | Authors: | Bordelon, T, Wilkinson, S.P, Grove, A, Newcomer, M.E. | | Deposit date: | 2005-12-09 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Transcriptional Regulator HucR from Deinococcus radiodurans Reveals a Repressor Preconfigured for DNA Binding.
J.Mol.Biol., 360, 2006
|
|
2FNQ
 
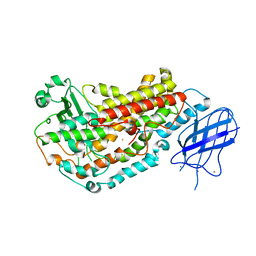 | |
2IDK
 
 | | Crystal Structure of Rat Glycine N-Methyltransferase Complexed With Folate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Glycine N-methyltransferase | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Egli, M, Newcomer, M.E, Wagner, C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 5-methyltetrahydrofolate is bound in intersubunit areas of rat liver folate-binding protein glycine N-methyltransferase.
J.Biol.Chem., 282, 2007
|
|
