5OBY
 
 | | Mycoplasma genitalium DnaK-NBD in complex with AMP-PNP | | Descriptor: | Chaperone protein DnaK, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5OBV
 
 | | Mycoplasma genitalium DnaK deletion mutant lacking SBDalpha in complex with ADP and Pi. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
6HVC
 
 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-Trp-(N-Me)Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2019-04-24 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
6HVB
 
 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-(N-Me)Trp-Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
6D1L
 
 | | Design, synthesis, and X-ray of selenides bearing benzenesulfonamide moiety with neuropathic pain modulating effects | | Descriptor: | 4-[(but-2-yn-1-yl)selanyl]benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Angeli, A, di Cesare Mannelli, L, Micheli, L, Ghelardini, C, Supuran, C.T. | | Deposit date: | 2018-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis and X-ray crystallography of selenides bearing benzenesulfonamide moiety with neuropathic pain modulating effects.
Eur J Med Chem, 154, 2018
|
|
6D1M
 
 | | Design, synthesis, and X-ray of selenides bearing benzenesulfonamide moiety with neuropathic pain modulating effects | | Descriptor: | 4-(cyclohexylselanyl)benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Angeli, A, di Cesare Mannelli, L, Micheli, L, Ghelardini, C, Supuran, C.T. | | Deposit date: | 2018-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design, synthesis and X-ray crystallography of selenides bearing benzenesulfonamide moiety with neuropathic pain modulating effects.
Eur J Med Chem, 154, 2018
|
|
6EBE
 
 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
1JWW
 
 | | NMR characterization of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
6CEH
 
 | | Design, Synthesis, X-ray and Biological Activities of Selenides Bearing the Benzenesulfonamide Moiety as New Class of Agents for Prevention of Diabetic Cerebrovascular Pathology | | Descriptor: | 4-[(prop-2-en-1-yl)selanyl]benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Angeli, A, di Cesare Mannelli, L, Trallori, E, Ghelardini, C, Carta, F, Supuran, C.T. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Design, Synthesis, and X-ray of Selenides as New Class of Agents for Prevention of Diabetic Cerebrovascular Pathology.
ACS Med Chem Lett, 9, 2018
|
|
1KQK
 
 | | Solution Structure of the N-terminal Domain of a Potential Copper-translocating P-type ATPase from Bacillus subtilis in the Cu(I)loaded State | | Descriptor: | COPPER (I) ION, POTENTIAL COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F.C, Ruiz-Duenas, F.J. | | Deposit date: | 2002-01-07 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
1SO9
 
 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
3D5O
 
 | | Structural recognition and functional activation of FcrR by innate pentraxins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low affinity immunoglobulin gamma Fc region receptor II-a, ... | | Authors: | Lu, J, Marnell, L.L, Marjon, K.D, Mold, C, Du Clos, T.W, Sun, P.D. | | Deposit date: | 2008-05-16 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural recognition and functional activation of FcgammaR by innate pentraxins.
Nature, 456, 2008
|
|
1RP5
 
 | | PBP2x from Streptococcus pneumoniae strain 5259 with reduced susceptibility to beta-lactam antibiotics | | Descriptor: | SULFATE ION, penicillin-binding protein 2x | | Authors: | Pernot, L, Chesnel, L, Legouellec, A, Croize, J, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics.
J.Biol.Chem., 279, 2004
|
|
1SP0
 
 | | Solution Structure of apoCox11 | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
3JB5
 
 | |
2GGP
 
 | | Solution structure of the Atx1-Cu(I)-Ccc2a complex | | Descriptor: | COPPER (I) ION, Metal homeostasis factor ATX1, Probable copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Cantini, F, Felli, I.C, Gonnelli, L, Hadjiliadis, N, Pierattelli, R, Rosato, A, Voulgaris, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction.
Nat.Chem.Biol., 2, 2006
|
|
2JT5
 
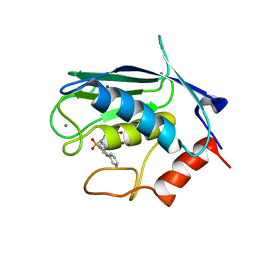 | | solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of n-hydroxy-2-[n-(2-hydroxyethyl)biphenyl-4-sulfonamide] hydroxamic acid (MLC88) | | Descriptor: | CALCIUM ION, N~2~-(biphenyl-4-ylsulfonyl)-N-hydroxy-N~2~-(2-hydroxyethyl)glycinamide, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-20 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JSD
 
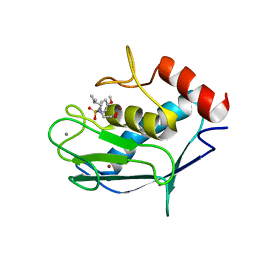 | | Solution structure of MMP20 complexed with NNGH | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-20, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Arendt, Y, Banci, L, Bertini, I, Cantini, F, Cozzi, R, Del Conte, R, Gonnelli, L, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-07-03 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Catalytic domain of MMP20 (Enamelysin) - the NMR structure of a new matrix metalloproteinase.
Febs Lett., 581, 2007
|
|
2JT6
 
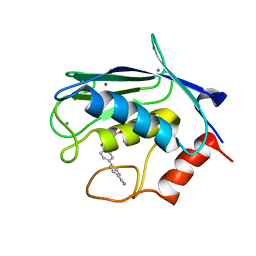 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of 3-4'-cyanobyphenyl-4-yloxy)-n-hdydroxypropionamide (MMP-3 inhibitor VII) | | Descriptor: | 3-[(4'-cyanobiphenyl-4-yl)oxy]-N-hydroxypropanamide, CALCIUM ION, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-23 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JNP
 
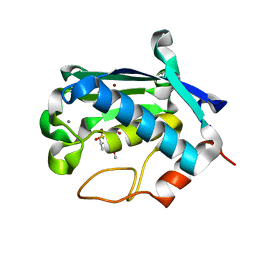 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-3, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-01-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
3HJH
 
 | | A rigid N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor | | Descriptor: | COBALT (II) ION, Transcription-repair-coupling factor | | Authors: | Murphy, M, Gong, P, Ralto, K, Manelyte, L, Savery, N, Theis, K. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd.
Nucleic Acids Res., 37, 2009
|
|
3ZTV
 
 | | Structure of Haemophilus influenzae NAD nucleotidase (NadN) | | Descriptor: | ADENOSINE, GLYCEROL, NAD NUCLEOTIDASE, ... | | Authors: | Garavaglia, S, Bruzzone, S, Cassani, C, Canella, L, Allegrone, G, Sturla, L, Mannino, E, Millo, E, De Flora, A, Rizzi, M. | | Deposit date: | 2011-07-12 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The High-Resolution Crystal Structure of Periplasmic Haemophilus Influenzae Nad Nucleotidase Reveals a Novel Enzymatic Function of Human Cd73 Related to Nad Metabolism.
Biochem.J., 441, 2012
|
|
3ZU0
 
 | | Structure of Haemophilus influenzae NAD nucleotidase (NadN) | | Descriptor: | NAD NUCLEOTIDASE, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Garavaglia, S, Bruzzone, S, Cassani, C, Canella, L, Allegrone, G, Sturla, L, Mannino, E, Millo, E, De Flora, A, Rizzi, M. | | Deposit date: | 2011-07-13 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The High-Resolution Crystal Structure of Periplasmic Haemophilus Influenzae Nad Nucleotidase Reveals a Novel Enzymatic Function of Human Cd73 Related to Nad Metabolism.
Biochem.J., 441, 2012
|
|
2LU5
 
 | | Structure and chemical shifts of Cu(I),Zn(II) superoxide dismutase by solid-state NMR | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn] | | Authors: | Knight, M.J, Pell, A.J, Bertini, I, Felli, I.C, Gonnelli, L, Pierattelli, R, Herrmann, T, Emsley, L, Pintacuda, G. | | Deposit date: | 2012-06-08 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KVY
 
 | | NMR solution structure of the 4:1 complex between an uncharged distamycin A analogue and [d(TGGGGT)]4 | | Descriptor: | 4-amino-1-methyl-N-{1-methyl-5-[(1-methyl-5-{[3-(methylamino)-3-oxopropyl]carbamoyl}-1H-pyrrol-3-yl)carbamoyl]-1H-pyrrol-3-yl}-1H-pyrrole-2-carboxamide, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3') | | Authors: | Cosconati, S, Marinelli, L, Trotta, R, Virno, A, De Tito, S, Romagnoli, R, Pagano, B, Limongelli, V, Giancola, C, Baraldi, P, Mayol, L, Novellino, E, Randazzo, A. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle.
J.Am.Chem.Soc., 132, 2010
|
|
