2QDN
 
 | |
3K51
 
 | | Crystal Structure of DcR3-TL1A complex | | Descriptor: | Decoy receptor 3, Tumor necrosis factor ligand superfamily member 15, secreted form | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2009-10-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3BP5
 
 | | Crystal structure of the mouse PD-1 and PD-L2 complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Yan, Q, Lazar-Molnar, E, Cao, E, Ramagopal, U.A, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-18 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BP6
 
 | | Crystal structure of the mouse PD-1 Mutant and PD-L2 complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Yan, Q, Lazar-Molnar, E, Cao, E, Ramagopal, U.A, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the mouse PD-1 A99L and PD-L2 complex
To be published
|
|
1NCN
 
 | |
4DB5
 
 | | Crystal structure of Rabbit GITRL | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tumor necrosis factor ligand superfamily member 18 | | Authors: | Kumar, P.R, Bhosle, R, Gizzi, A, Scott Glenn, A, Chowhury, S, Hillerich, B, Seidel, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.522 Å) | | Cite: | Crystal structure of GITRL from Oryctolagus cuniculus
to be published
|
|
2IF7
 
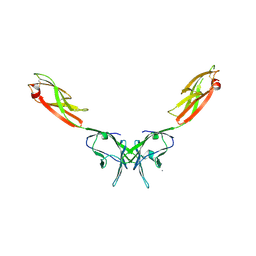 | | Crystal Structure of NTB-A | | Descriptor: | CALCIUM ION, CHLORIDE ION, SLAM family member 6 | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-17 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | NTB-A Receptor Crystal Structure: Insights into Homophilic Interactions in the Signaling Lymphocytic Activation Molecule Receptor Family.
Immunity, 25, 2006
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
3B9I
 
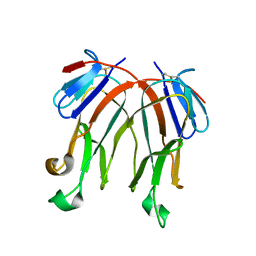 | |
1DQT
 
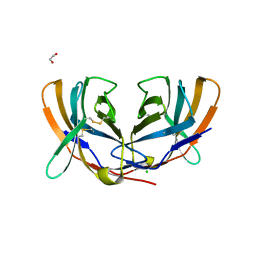 | | THE CRYSTAL STRUCTURE OF MURINE CTLA4 (CD152) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTOTOXIC T LYMPHOCYTE ASSOCIATED ANTIGEN 4 | | Authors: | Ostrov, D.A, Shi, W, Schwartz, J.C, Almo, S.C, Nathenson, S.G. | | Deposit date: | 2000-01-05 | | Release date: | 2000-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of murine CTLA-4 and its role in modulating T cell responsiveness.
Science, 290, 2000
|
|
3MHD
 
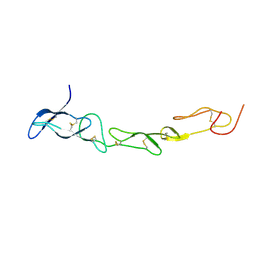 | | Crystal structure of DCR3 | | Descriptor: | Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3MI8
 
 | | The structure of TL1A-DCR3 COMPLEX | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 15, SECRETED FORM, Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-09 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3NWM
 
 | | Crystal structure of a single chain construct composed of MHC class I H-2Kd, beta-2microglobulin and a peptide which is an autoantigen for type 1 diabetes | | Descriptor: | Peptide/beta-2microglobulin/MHC class I H-2Kd chimeric protein | | Authors: | Ramagopal, U.A, Samanta, D, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of a single-chain peptide-MHC molecule that modulates both naive and activated CD8+ T cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2MHA
 
 | | CRYSTAL STRUCTURE OF THE MAJOR HISTOCOMPATIBILITY COMPLEX CLASS I H-2KB MOLECULE CONTAINING A SINGLE VIRAL PEPTIDE: IMPLICATIONS FOR PEPTIDE BINDING AND T-CELL RECEPTOR RECOGNITION | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (H-2KB) (ALPHA CHAIN), VIRAL OCTAPEPTIDE ARG-GLY-TYR-VAL-TYR-GLN-GLY-LEU | | Authors: | Zhang, W, Young, A.C.M, Imarai, M, Nathenson, S.G, Sacchettini, J.C. | | Deposit date: | 1993-07-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the major histocompatibility complex class I H-2Kb molecule containing a single viral peptide: implications for peptide binding and T-cell receptor recognition.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
2OYP
 
 | | T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface | | Descriptor: | Hepatitis A virus cellular receptor 2, SULFATE ION | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Immunity, 26, 2007
|
|
2Q1M
 
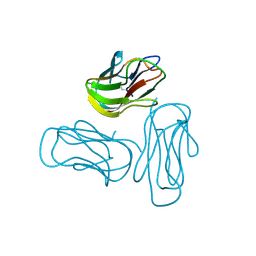 | |
2PKD
 
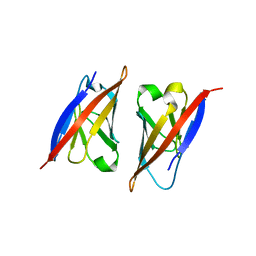 | | Crystal structure of CD84: Insite into SLAM family function | | Descriptor: | CHLORIDE ION, SLAM family member 5 | | Authors: | Yan, Q, Malashkevich, V.N, Fedorov, A, Cao, E, Lary, J.W, Cole, J.L, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-04-17 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structure of CD84 provides insight into SLAM family function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1INQ
 
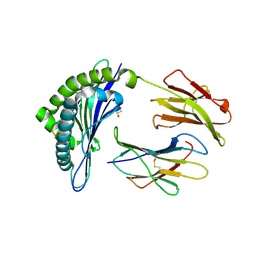 | | Structure of Minor Histocompatibility Antigen peptide, H13a, complexed to H2-Db | | Descriptor: | BETA-2 MICROGLOBULIN, DIMETHYL SULFOXIDE, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D, Nathenson, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
4J6G
 
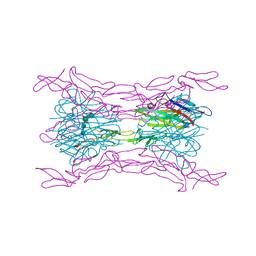 | | CRYSTAL STRUCTURE OF LIGHT AND DcR3 COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-11 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
1I85
 
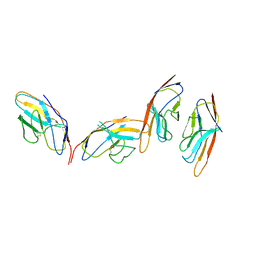 | | CRYSTAL STRUCTURE OF THE CTLA-4/B7-2 COMPLEX | | Descriptor: | CYTOTOXIC T-LYMPHOCYTE-ASSOCIATED PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD86 | | Authors: | Schwartz, J.-C.D, Zhang, X, Fedorov, A.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2001-03-12 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for co-stimulation by the human CTLA-4/B7-2 complex.
Nature, 410, 2001
|
|
4JJH
 
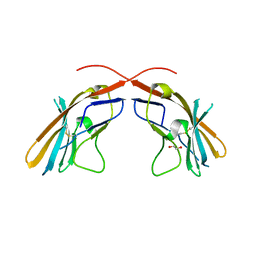 | | Crystal structure of the D1 domain from human Nectin-4 extracellular fragment [PSI-NYSGRC-005624] | | Descriptor: | 1,2-ETHANEDIOL, Poliovirus receptor-related protein 4 | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D1 domain of human Nectin-4 extracellular fragment [NYSGRC-005624]
to be published
|
|
4JGJ
 
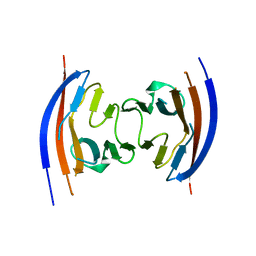 | | Crystal structure of the Ig-like D1 domain from mouse Carcinoembryogenic antigen-related cell adhesion molecule 15 (CEACAM15) [PSI-NYSGRC-005691] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 15, Unknown peptide | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6508 Å) | | Cite: | Crystal structure of the Ig-like D1 domain of CEACAM15 from Mus musculus [NYSGRC-005691]
to be published
|
|
4KG8
 
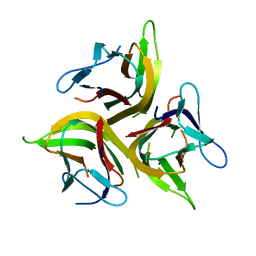 | | Crystal structure of light mutant | | Descriptor: | Tumor necrosis factor ligand superfamily member 14 | | Authors: | Liu, W, Zhan, C, Kumar, P.R, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-28 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4KGQ
 
 | | Crystal structure of a human light loop mutant in complex with dcr3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Sampathkumar, P, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-29 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
1LD9
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF AN H-2LD PEPTIDE COMPLEX EXPLAINS THE UNIQUE INTERACTION OF LD WITH BETA2M AND PEPTIDE | | Descriptor: | BETA-2 MICROGLOBULIN, MHC CLASS I H-2LD HEAVY CHAIN, NANO-PEPTIDE | | Authors: | Balendiran, G.K, Solheim, J.C, Young, A.C.M, Hansen, T.H, Nathenson, S.G, Sacchettini, J.C. | | Deposit date: | 1997-04-24 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of an H-2Ld-peptide complex explains the unique interaction of Ld with beta-2 microglobulin and peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
