2MW5
 
 | |
2MOT
 
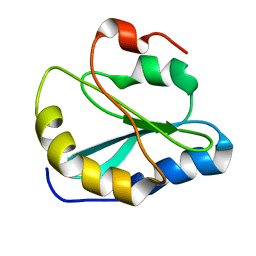 | | Backbone Structure of Actin Depolymerizing Factor (ADF) of Toxoplasma gondii Based on Prot3DNMR Approach | | Descriptor: | Actin depolymerizing factor ADF | | Authors: | Kumar, D, Raikwal, N, Raval, I, Jaiswal, N, Shukla, V, Arora, A. | | Deposit date: | 2014-05-05 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Prot3DNMR: A Simple and Swift Strategy for Backbone Structure Determination of Proteins by NMR
To be Published
|
|
7BGI
 
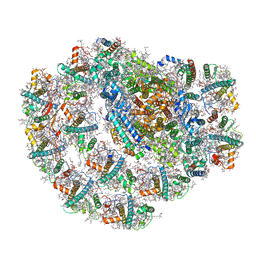 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Nelson, N. | | Deposit date: | 2021-01-07 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
7BLX
 
 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Nelson, N. | | Deposit date: | 2021-01-19 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
1UJ0
 
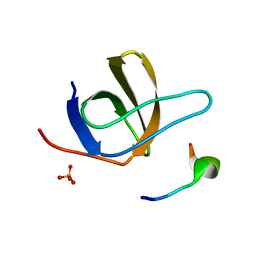 | | Crystal Structure of STAM2 SH3 domain in complex with a UBPY-derived peptide | | Descriptor: | PHOSPHATE ION, deubiquitinating enzyme UBPY, signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | | Authors: | Kaneko, T, Kumasaka, T, Ganbe, T, Sato, T, Miyazawa, K, Kitamura, N, Tanaka, N. | | Deposit date: | 2003-07-24 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain.
J.Biol.Chem., 278, 2003
|
|
1VAJ
 
 | | Crystal Structure of Uncharacterized Protein PH0010 From Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0010 | | Authors: | Tajika, Y, Sakai, N, Tamura, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-02-17 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of PH0010 from Pyrococcus horikoshii, which is highly homologous to human AMMECR 1C-terminal region
Proteins, 58, 2005
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
1RJD
 
 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | BETA-MERCAPTOETHANOL, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity
J.Biol.Chem., 279, 2004
|
|
5M1H
 
 | |
1RJE
 
 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | BETA-MERCAPTOETHANOL, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, de La Sierra-Gallay, I.L, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity.
J.Biol.Chem., 279, 2004
|
|
1V38
 
 | | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1 | | Descriptor: | SAM-domain protein SAMSN-1 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-29 | | Release date: | 2004-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1
To be Published
|
|
7X7O
 
 | | SARS-CoV-2 spike RBD in complex with neutralizing antibody UT28K | | Descriptor: | Spike protein S1, UT28K Fab, heavy chain, ... | | Authors: | Ozawa, T, Tani, H, Anraku, Y, Kita, S, Igarashi, E, Saga, Y, Inasaki, N, Kawasuji, H, Yamada, H, Sasaki, S, Somekawa, M, Sasaki, J, Hayakawa, Y, Yamamoto, Y, Morinaga, Y, Kurosawa, N, Isobe, M, Fukuhara, H, Maenaka, K, Hashiguchi, T, Kishi, H, Kitajima, I, Saito, S, Niimi, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Novel super-neutralizing antibody UT28K is capable of protecting against infection from a wide variety of SARS-CoV-2 variants.
Mabs, 14, 2022
|
|
2L5I
 
 | | structure of the spliceosomal phosphopeptide P140 (non-phosphorylated form) | | Descriptor: | U1 small nuclear ribonucleoprotein 70 kDa | | Authors: | Quinternet, M, Page, N, Schall, N, Strub, J, Chaloin, O, Decossas, M, Cung, M, van Dorsselaer, A, Briand, J, Muller, S. | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The spliceosomal phosphopeptide P140 controls the lupus disease by interacting with the HSC70 protein and via a mechanism mediated by gammadelta T cells.
Plos One, 4, 2009
|
|
1V7Z
 
 | | creatininase-product complex | | Descriptor: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-12-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
7W8G
 
 | | Cryo-EM structure of MCM double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Cheng, J, Li, N, Tye, B, Zhai, Y, Gao, N. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
2OWK
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, diphthine synthase | | Authors: | Sugahara, M, Matsuura, Y, Ono, N, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-16 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
4CVD
 
 | |
1V40
 
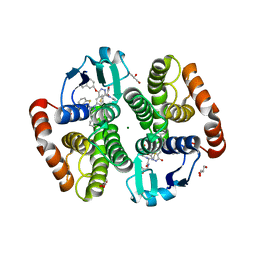 | | First Inhibitor Complex Structure of Human Hematopoietic Prostaglandin D Synthase | | Descriptor: | 3-(1,3-BENZOTHIAZOL-2-YL)-2-(1,4-DIOXO-1,2,3,4-TETRAHYDROPHTHALAZIN-6-YL)-5-[(E)-2-PHENYLVINYL]-3H-TETRAAZOL-2-IUM, GLUTATHIONE, GLYCEROL, ... | | Authors: | Inoue, T, Okano, Y, Kado, Y, Aritake, K, Irikura, D, Uodome, N, Kinugasa, S, Okazaki, N, Matsumura, H, Kai, Y, Urade, Y. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First determination of the inhibitor complex structure of human hematopoietic prostaglandin D synthase.
J.Biochem.(Tokyo), 135, 2004
|
|
1V85
 
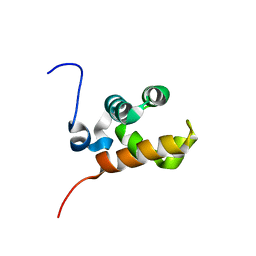 | | Sterile alpha motif (SAM) domain of mouse bifunctional apoptosis regulator | | Descriptor: | similar to ring finger protein 36 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Hayashi, F, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-29 | | Release date: | 2005-01-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sterile alpha motif (SAM) domain of mouse bifunctional apoptosis regulator
To be Published
|
|
2P5C
 
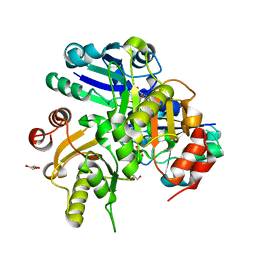 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
1V4N
 
 | | Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase homologue from Sulfolobus tokodaii | | Descriptor: | 271aa long hypothetical 5'-methylthioadenosine phosphorylase | | Authors: | Kitago, Y, Yasutake, Y, Sakai, N, Tsujimura, M, Yao, M, Watanabe, N, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2003-11-14 | | Release date: | 2005-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii MTAP
To be Published
|
|
1V30
 
 | | Crystal Structure Of Uncharacterized Protein PH0828 From Pyrococcus horikoshii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hypothetical UPF0131 protein PH0828 | | Authors: | Tajika, Y, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-10-21 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of hypothetical protein PH0828 from Pyrococcus horikoshii.
Proteins, 57, 2004
|
|
2PCH
 
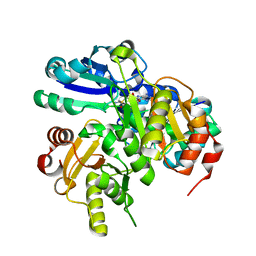 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | Authors: | Sugahara, M, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2L5J
 
 | | structure of the spliceosomal phosphopeptide P140 (phosphorylated form) | | Descriptor: | U1 small nuclear ribonucleoprotein 70 kDa | | Authors: | Quinternet, M, Page, N, Schall, N, Strub, J, Chaloin, O, Decossas, M, Cung, M, van Dorsselaer, A, Briand, J, Muller, S. | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The spliceosomal phosphopeptide P140 controls the lupus disease by interacting with the HSC70 protein and via a mechanism mediated by gammadelta T cells.
Plos One, 4, 2009
|
|
2OUM
 
 | | The first domain of L1 from Thermus thermophilus | | Descriptor: | 50S ribosomal protein L1 | | Authors: | Kljashtorny, V, Tishchenko, S, Nevskaya, N, Nikonov, S, Davydova, N, Garber, M. | | Deposit date: | 2007-02-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Domain I of ribosomal protein L1 is sufficient for specific RNA binding.
Nucleic Acids Res., 35, 2007
|
|
