6VWL
 
 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P/E tRNA (rotated conformation) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
6VZV
 
 | | TTLL6 bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(3~{S})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W1T
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the second illumination at 2.01 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1S5Z
 
 | | NDP kinase in complex with adenosine phosphonoacetic acid | | Descriptor: | ADENOSINE PHOSPHONOACETIC ACID, Nucleoside diphosphate kinase, cytosolic, ... | | Authors: | Chen, Y, Morera, S, Pasti, C, Angusti, A, Solaroli, N, Veron, M, Janin, J, Manfredini, S, Deville-Bonne, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Adenosine phosphonoacetic acid is slowly metabolized by NDP kinase.
Med.Chem., 1, 2005
|
|
1S8D
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-3A | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-02-02 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
6VWN
 
 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P-site tRNA (non-rotated conformation, Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
1SCZ
 
 | | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase | | Descriptor: | Dihydrolipoamide Succinyltransferase | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Tsao, J, Johnson, D, Huang, W.-Y, Pruett, P, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, Z, Lu, S, DeLucas, L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase
To be Published
|
|
3ATZ
 
 | | Crystal structure of TcOYE with pHBA | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE, Prostaglandin F2a synthase | | Authors: | Okamoto, N, Yamaguchi, K, Mizohata, E, Tokuoka, K, Uchiyama, N, Sugiyama, S, Matsumura, H, Inaka, K, Urade, Y, Inoue, T. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insight into the stereoselective production of PGF2(alpha) by Old Yellow Enzyme from Trypanosoma cruzi
J.Biochem., 150, 2011
|
|
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | Descriptor: | Non-structural protein 9 | | Authors: | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
1SJ0
 
 | | Human Estrogen Receptor Alpha Ligand-binding Domain in Complex with the Antagonist Ligand 4-D | | Descriptor: | (2S,3R)-2-(4-(2-(PIPERIDIN-1-YL)ETHOXY)PHENYL)-2,3-DIHYDRO-3-(4-HYDROXYPHENYL)BENZO[B][1,4]OXATHIIN-6-OL, Estrogen receptor | | Authors: | Kim, S, Wu, J.Y, Birzin, E.T, Chan, W, Pai, L.Y, Yang, Y.T, Mosley, R.T, Fitzgerald, P.M, Sharma, N, DiNinno, F, Rohrer, S.P, Schaeffer, J.M, Hammond, M.L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estrogen Receptor Ligands. II. Discovery of Benzoxathiins as Potent, Selective Estrogen Receptor alpha Modulators.
J.Med.Chem., 47, 2004
|
|
6W5B
 
 | |
1SNU
 
 | | CRYSTAL STRUCTURE OF THE UNPHOSPHORYLATED INTERLEUKIN-2 TYROSINE KINASE CATALYTIC DOMAIN | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
6WVH
 
 | | Human VKOR with Brodifacoum | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Brodifacoum, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
1S9B
 
 | | Crystal Structure Analysis of the B-DNA GAATTCG | | Descriptor: | 5'-D(*GP*AP*AP*TP*TP*CP*G)-3', NICKEL (II) ION | | Authors: | Valls, N, Uson, I, Gouyette, C, Subirana, J.A. | | Deposit date: | 2004-02-04 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A cubic arrangement of DNA double helices based on nickel-guanine interactions
J.Am.Chem.Soc., 126, 2004
|
|
6WEZ
 
 | | Crystal Structure of Broadly Neutralizing Antibody 3I14-D93N Mutant Bound to the Influenza A H3 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hemagglutinin, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
1SJQ
 
 | | NMR Structure of RRM1 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
1SIO
 
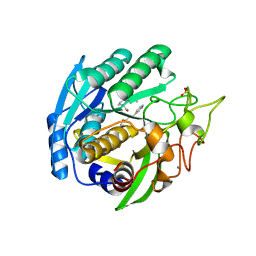 | | Structure of Kumamolisin-As complexed with a covalently-bound inhibitor, AcIPF | | Descriptor: | Ace-ILE-PRO-PHL peptide inhibitor, CALCIUM ION, SULFATE ION, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Tsuruoka, N, Ashida, M, Minakata, H, Oyama, H, Oda, K, Nishino, T, Nakayama, T. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity
J.Biol.Chem., 279, 2004
|
|
6WNT
 
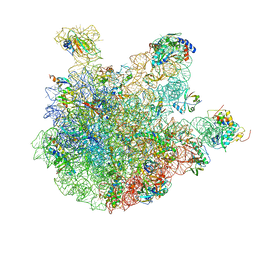 | | 50S ribosomal subunit without free 5S rRNA and perturbed PTC | | Descriptor: | 23s-5s joint ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
1SNL
 
 | |
6VJG
 
 | | Csx3-I222 Crystal Form at 1.8 Angstrom Resolution | | Descriptor: | CRISPR-associated protein, Csx3 family | | Authors: | Brown, S, Charbonneau, A, Burman, N, Gauvin, C.C, Lawrence, C.M. | | Deposit date: | 2020-01-15 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA 4 .
J.Biol.Chem., 295, 2020
|
|
1SQY
 
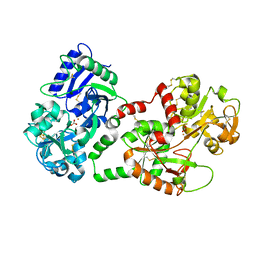 | | Structure of human diferric lactoferrin at 2.5A resolution using crystals grown at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Vikram, P, Prem Kumar, R, Singh, N, Kumar, J, Ethayathulla, A.S, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of human diferric lactoferrin at 2.5A resolution using crystals grown at pH 6.5.
To be Published
|
|
1SG7
 
 | | NMR solution structure of the putative cation transport regulator ChaB | | Descriptor: | Putative Cation transport regulator chaB | | Authors: | Osborne, M.J, Siddiqui, N, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-23 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ChaB, a putative membrane ion antiporter regulator from Escherichia coli
BMC STRUCT.BIOL., 4, 2004
|
|
1SH3
 
 | | Crystal Structure of Norwalk Virus Polymerase (MgSO4 crystal form) | | Descriptor: | MAGNESIUM ION, RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
6VRC
 
 | | Cryo-EM structure of Cas13(crRNA) | | Descriptor: | CRISPR-associated endoribonuclease Cas13a, RNA (51-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
1RXA
 
 | |
