4RXZ
 
 | |
2RUW
 
 | | Solution structures of the DNA-binding domain (ZF5) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
8T21
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein | | Descriptor: | Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
2RV1
 
 | | Solution structures of the DNA-binding domain (ZF13) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
5HCT
 
 | | Endothiapepsin in complex with biacylhydrazone | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-N'-{3-[(E)-{2-[(2S)-2-amino-3-(1H-indol-3-yl)propanoyl]hydrazinylidene}methyl]benzylidene}-3-(1H-indol-2-yl)propanehydrazide (non-preferred name), ACETATE ION, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2016-01-04 | | Release date: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fragment Linking and Optimization of Inhibitors of the Aspartic Protease Endothiapepsin: Fragment-Based Drug Design Facilitated by Dynamic Combinatorial Chemistry.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5A39
 
 | | Structure of Rad14 in complex with cisplatin containing DNA | | Descriptor: | 5'-D(*DGP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP*GP)-3', 5'-D(*DTP*GP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP)-3', Cisplatin, ... | | Authors: | Koch, S.C, Kuper, J, Gasteiger, K.L, Wichlein, N, Strasser, R, Eisen, D, Geiger, S, Schneider, S, Kisker, C, Carell, T. | | Deposit date: | 2015-05-28 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Recognition of Cisplatin and Aaf-Dg Lesion by Rad14 (Xpa).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8T22
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
4TM7
 
 | | Crystal structure of 6-phosphogluconolactonase from Mycobacterium smegmatis N131D mutant soaked with CuSO4 | | Descriptor: | 1,2-ETHANEDIOL, 6-phosphogluconolactonase, CHLORIDE ION, ... | | Authors: | Fujieda, N, Stuttfeld, E, Maier, T. | | Deposit date: | 2014-05-31 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Enzyme repurposing of a hydrolase as an emergent peroxidase upon metal binding.
Chem Sci, 6, 2015
|
|
4TMB
 
 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
8T25
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at downRBD conformation. | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T20
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
2RUV
 
 | | Solution structures of the DNA-binding domain (ZF4) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
8T23
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
2RJF
 
 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 12-30), orthorhombic form I | | Descriptor: | Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RV5
 
 | | Solution structures of the DNA-binding domain (ZF8) of mouse immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
4RKO
 
 | | Crystal structure of thrombin mutant S195T bound with PPACK | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | Deposit date: | 2014-10-13 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
2RNL
 
 | | Solution structure of the EGF-like domain from human Amphiregulin | | Descriptor: | Amphiregulin | | Authors: | Qin, X, Hayashi, F, Terada, T, Shirouzu, M, Watanabe, S, Kigawa, T, Yabuta, N, Nojima, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the EGF-like domain from human Amphiregulin
To be Published
|
|
4RN5
 
 | | B1 domain of human Neuropilin-1 with acetate ion in a ligand-binding site | | Descriptor: | ACETATE ION, GLYCEROL, Neuropilin-1, ... | | Authors: | Allerston, C.K, Yelland, T.S, Jarvis, A, Jenkins, K, Winfield, N, Cheng, L, Jia, H, Zachary, I, Selwood, D.L, Djordjevic, S. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Conserved water molecules in a ligand-binding site of neuropilin-1
To be Published
|
|
4ROL
 
 | | Deoxyhemoglobin in complex with imidazolylacryloyl derivatives | | Descriptor: | 4-{2,3-dichloro-4-[3-(1H-imidazol-2-yl)propanoyl]phenoxy}butanoic acid, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Omar, A.M, Mahran, M.A, Ghatge, M.N, Chowdhury, N, Bamane, F.H.A, El-Araby, M.E, Abdulmalik, O, Safo, M.K. | | Deposit date: | 2014-10-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a novel class of covalent modifiers of hemoglobin as potential antisickling agents.
Org.Biomol.Chem., 13, 2015
|
|
2RV3
 
 | | Solution structures of the DNA-binding domain (ZF15) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RSJ
 
 | |
2RJE
 
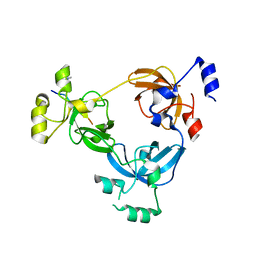 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), orthorhombic form II | | Descriptor: | CHLORIDE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
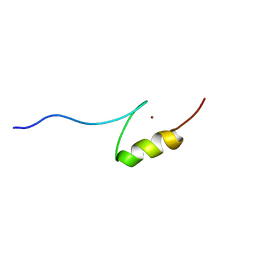
2RSH
 
 | |
8PEK
 
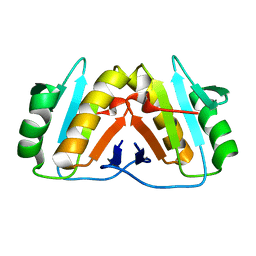 | |
8P9R
 
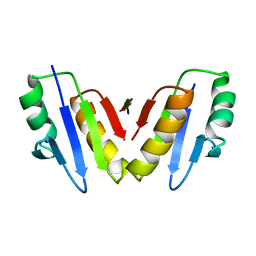 | | Structure of the periplasmic domain of ExbD from E. coli in complex with TonB | | Descriptor: | Biopolymer transport protein ExbD, Protein TonB | | Authors: | Zinke, M, Mechaly, A, Lejeune, M, Haouz, A, Izadi-Pruneyre, N. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ton motor conformational switch and peptidoglycan role in bacterial nutrient uptake.
Nat Commun, 15, 2024
|
|
