2E6I
 
 | | Solution structure of the BTK motif of tyrosine-protein kinase ITK from human | | Descriptor: | Tyrosine-protein kinase ITK/TSK, ZINC ION | | Authors: | Li, H, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-27 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the BTK motif of tyrosine-protein kinase ITK from human
To be Published
|
|
5XLS
 
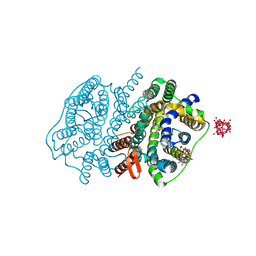 | | Crystal structure of UraA in occluded conformation | | Descriptor: | 12-TUNGSTOPHOSPHATE, URACIL, Uracil permease | | Authors: | Yu, X.Z, Yang, G.H, Yan, C.Y, Yan, N. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters
Cell Res., 27, 2017
|
|
6RXY
 
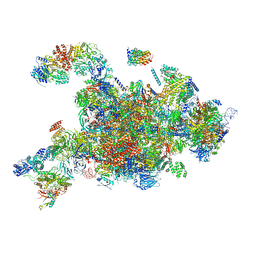 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state a | | Descriptor: | 35S rRNA, 40S ribosomal protein S13-like protein, 40S ribosomal protein S14-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6S75
 
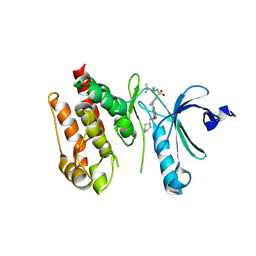 | | Crystal structure of Nek7 bound to compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, Serine/threonine-protein kinase Nek7 | | Authors: | Nasir, N, Bayliss, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
7K3D
 
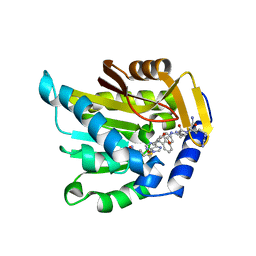 | | The structure of NTMT1 in complex with compound DC1-13 | | Descriptor: | N-terminal Xaa-Pro-Lys N-methyltransferase 1, N~2~-{(2S)-1-[(naphthalen-1-yl)acetyl]-2,5-dihydro-1H-pyrrole-2-carbonyl}-L-lysyl-L-argininamide, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Chen, D, Huang, R, Noinaj, N. | | Deposit date: | 2020-09-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based Discovery of Cell-Potent Peptidomimetic Inhibitors for Protein N-Terminal Methyltransferase 1.
Acs Med.Chem.Lett., 12, 2021
|
|
2E9K
 
 | | Solution structure of the CH domain from human MICAL-2 | | Descriptor: | Protein MICAL-2 | | Authors: | Tomizawa, T, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from human MICAL-2
To be Published
|
|
6S21
 
 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33494S-sulf) | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Endo-4-O-sulfatase | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
2EDB
 
 | |
5XN0
 
 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5EZR
 
 | | Crystal Structure of PVX_084705 bound to compound | | Descriptor: | CHLORIDE ION, N-[5-(3-{2-[(cyclopropylmethyl)amino]pyrimidin-4-yl}-7-[(dimethylamino)methyl]-6-methylimidazo[1,2-a]pyridin-2-yl)-2-fluorophenyl]methanesulfonamide, cGMP-dependent protein kinase, ... | | Authors: | El Bakkouri, M, Amani, M, Walker, J.R, Osborne, S, Large, J.M, Birchall, K, Bouloc, N, Smiljanic-Hurley, E, Wheldon, M, Harding, D.J, Merritt, A.T, Ansell, K.H, Coombs, P.J, Kettleborough, C.A, Stewart, B.L, Bowyer, P.W, Gutteridge, W.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Baker, D.A, Hui, R, Loppnau, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-26 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PVX_084705 bound to compound
To Be Published
|
|
5XTY
 
 | |
5F0E
 
 | | Murine endoplasmic reticulum alpha-glucosidase II | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2015-11-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7ZJP
 
 | | Optimization of TEAD P-Site Binding Fragment Hit into In Vivo Active Lead MSC-4106 | | Descriptor: | 2-methyl-4-[4-(trifluoromethyl)phenyl]pyrazolo[3,4-b]indole-7-carboxylic acid, SULFATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Freire, F, Heinrich, T, Petersson, C, Schneider, R, Garg, S, Schwarz, D, Gunera, J, Seshire, A, Koetzner, L, Schlesiger, S, Musil, D, Schilke, H, Doerfel, B, Diehl, P, Boepple, P, Lemos, A.R, Sousa, P.M.F, Freire, F, Bandeiras, T.M, Carswell, E, Pearson, N, Sirohi, S, Hooker, M, Trivier, E, Broome, R, Balsiger, A, Crowden, A, Dillon, C, Wienke, D. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of TEAD P-Site Binding Fragment Hit into In Vivo Active Lead MSC-4106 .
J.Med.Chem., 65, 2022
|
|
2EL8
 
 | | Solution structure of the human STAP2 SH2 domain | | Descriptor: | Signal-transducing adaptor protein 2 | | Authors: | Kasai, T, Miyamoto, K, Tochio, N, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human STAP2 SH2 domain
To be Published
|
|
5F07
 
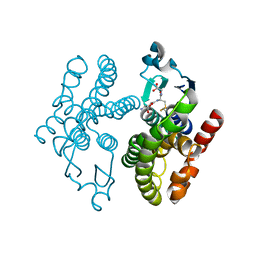 | | Crystal structure of glutathione transferase F8 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase family protein | | Authors: | Didierjean, C, Rouhier, N, Pegeot, H, Gense, F. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural plasticity among glutathione transferase Phi members: natural combination of catalytic residues confers dual biochemical activities.
FEBS J., 284, 2017
|
|
8A10
 
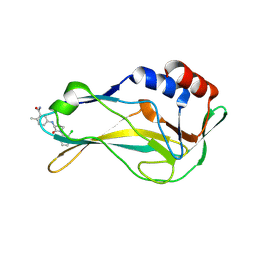 | | Crystal Structure of human Brachyury G177D variant in complex with Molpolrt-020-049-143 | | Descriptor: | T-box transcription factor T, UNKNOWN LIGAND | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-05-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with Molpolrt-020-049-143
To Be Published
|
|
5XXB
 
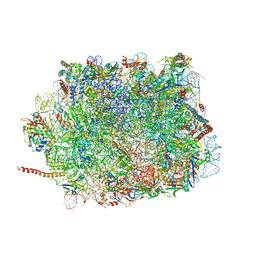 | | Large subunit of Toxoplasma gondii ribosome | | Descriptor: | 25S RNA, 5.8S RNA, 5S RNA, ... | | Authors: | Li, Z, Guo, Q, Zheng, L, Ji, Y, Xie, Y, Lai, D, Lun, Z, Suo, X, Gao, N. | | Deposit date: | 2017-07-03 | | Release date: | 2017-08-30 | | Last modified: | 2017-10-18 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii
Cell Res., 27, 2017
|
|
6S2L
 
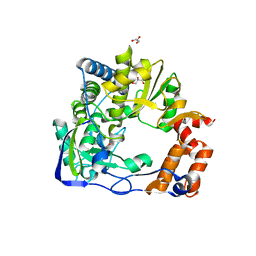 | |
5F5U
 
 | | Crystal structure of the Snu23-Prp38-MFAP1(217-258) complex of Chaetomium thermophilum | | Descriptor: | Prp38, Putative uncharacterized protein, Zinc finger domain-containing protein | | Authors: | Ulrich, A.K.C, Seeger, M, Bartlick, N, Wahl, M.C. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | Scaffolding in the Spliceosome via Single alpha Helices.
Structure, 24, 2016
|
|
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
6S3A
 
 | | Coxsackie B3 2C protein in complex with S-Fluoxetine | | Descriptor: | (3S)-N-methyl-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, 2C protein, CHLORIDE ION, ... | | Authors: | El Kazzi, P, Papageorgiou, N, Ferron, F.P, Bauer, L, van Kuppeveld, F, Coutard, B. | | Deposit date: | 2019-06-24 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fluoxetine targets an allosteric site in the enterovirus 2C AAA+ ATPase and stabilizes a ring-shaped hexameric complex.
Sci Adv, 8, 2022
|
|
5XN8
 
 | | Structure of glycerol dehydrogenase crystallised as a contaminant | | Descriptor: | GLYCEROL, Glycerol Dehydrogenase, ZINC ION | | Authors: | Hatti, K, Mathiharan, Y.K, Srinivasan, N, Murthy, M.R.N. | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Seeing but not believing: the structure of glycerol dehydrogenase initially assumed to be the structure of a survival protein from Salmonella typhimurium
Acta Crystallogr.,Sect.D, 73, 2017
|
|
2ECZ
 
 | | Solution structure of the SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Abe, H, Miyamoto, K, Tochio, N, Saito, K, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of Sorbin and SH3 domain-containing protein 1
To be Published
|
|
2EDD
 
 | | Solution structure of the fifth fibronectin type III domain of human Netrin receptor DCC | | Descriptor: | Netrin receptor DCC | | Authors: | Tochio, N, Koshiba, S, Tomizawa, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fifth fibronectin type III domain of human Netrin receptor DCC
To be Published
|
|
5XTA
 
 | | Crystal structure of lpg1832, a VirK family protein from Legionella pneumophila | | Descriptor: | GLYCEROL, SULFATE ION, VirK protein | | Authors: | Yin, S, Gong, X, Zhang, N, Ge, H. | | Deposit date: | 2017-06-18 | | Release date: | 2017-08-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lpg1832, a VirK family protein from Legionella pneumophila, reveals a novel fold for bacterial VirK proteins
FEBS Lett., 591, 2017
|
|
