7UPV
 
 | |
7THX
 
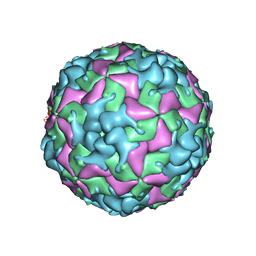 | | Cryo-EM structure of W6 possum enterovirus | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, I, Jayawardena, N, Strauss, M, Bostina, M. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM Structure of a Possum Enterovirus.
Viruses, 14, 2022
|
|
7TQW
 
 | | Kod RSGA incorporating PMT, n+2 | | Descriptor: | DNA polymerase, Primer, Template | | Authors: | Hajjar, M, Chim, N, Chaput, J.C. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystallographic analysis of engineered polymerases synthesizing phosphonomethylthreosyl nucleic acid.
Nucleic Acids Res., 50, 2022
|
|
7UUY
 
 | | Structure of the sodium/iodide symporter (NIS) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Sodium/iodide cotransporter | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
7UUZ
 
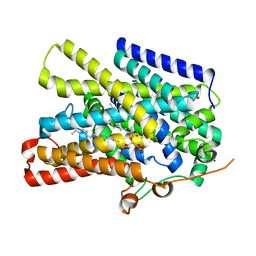 | | Structure of the sodium/iodide symporter (NIS) in complex with perrhenate and sodium | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, PERRHENATE, SODIUM ION, ... | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
7UV0
 
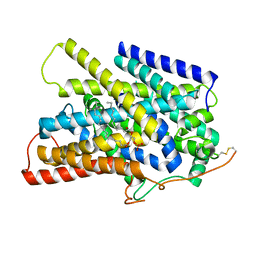 | | Structure of the sodium/iodide symporter (NIS) in complex with iodide and sodium | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, IODIDE ION, SODIUM ION, ... | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
3A5P
 
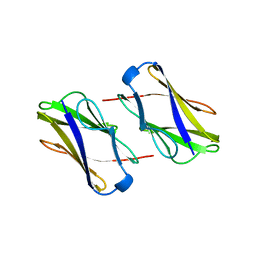 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
6UK1
 
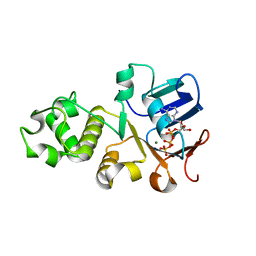 | | Crystal structure of nucleotide-binding domain 2 (NBD2) of the human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Vorobiev, S.M, Vernon, R.M, Khazanov, N, Senderowitz, H, Forman-Kay, J.D, Hunt, J.F. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | A thermodynamically stabilized form of the second nucleotide binding domain from human CFTR shows a catalytically inactive conformation
To Be Published
|
|
6UJI
 
 | |
1PGY
 
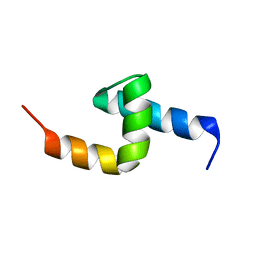 | | Solution structure of the UBA domain in Saccharomyces cerevisiae protein, Swa2p | | Descriptor: | Swa2p | | Authors: | Chim, N, Gall, W.E, Xiao, J, Harris, M.P, Graham, T.R, Krezel, A.M. | | Deposit date: | 2003-05-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-binding domain in Swa2p from Saccharomyces cerevisiae.
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
6UMR
 
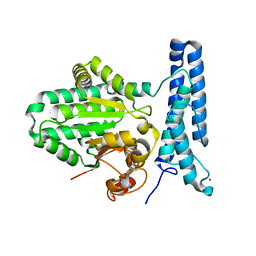 | | Structure of DUF89 - D291A mutant | | Descriptor: | Damage-control phosphatase DUF89, MAGNESIUM ION | | Authors: | Perry, J.J, Kenjic, N, Dennis, T.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Human ARMT1 structure and substrate specificity indicates that it is a DUF89 family damage-control phosphatase.
J.Struct.Biol., 212, 2020
|
|
1PC2
 
 | |
1ZN1
 
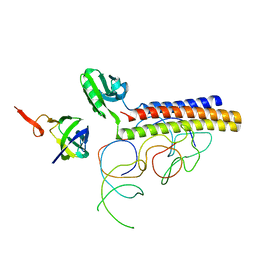 | | Coordinates of RRF fitted into Cryo-EM map of the 70S post-termination complex | | Descriptor: | 30S ribosomal protein S12, Ribosome recycling factor, ribosomal 16S RNA, ... | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
1PHW
 
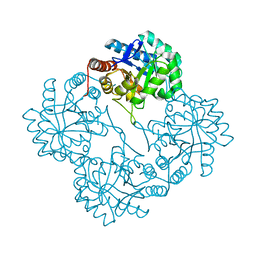 | | Crystal structure of KDO8P synthase in its binary complex with substrate analog 1-deoxy-A5P | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE | | Authors: | Vainer, R, Belakhov, V, Rabkin, E, Baasov, T, Adir, N. | | Deposit date: | 2003-05-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of Escherichia coli KDO8P synthase complexes reveal the source of catalytic irreversibility.
J.Mol.Biol., 351, 2005
|
|
1PHQ
 
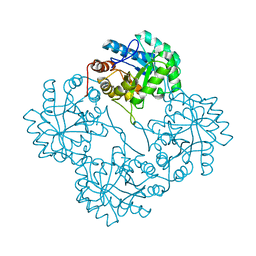 | | Crystal structure of KDO8P synthase in its binary complex with substrate analog E-FPEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, 3-FLUORO-2-(PHOSPHONOOXY)PROPANOIC ACID | | Authors: | Vainer, R, Adir, N, Baasov, T, Belakhov, V, Rabkin, E. | | Deposit date: | 2003-05-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic Analysis of the Phosphoenol Pyruvate Binding Site in E. Coli KDO8P Synthase
To be Published
|
|
6UD0
 
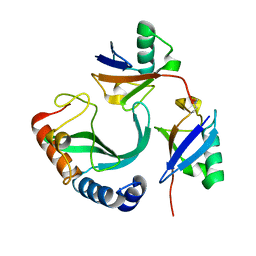 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
1PCO
 
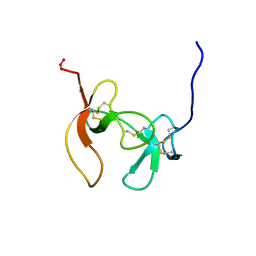 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PROCOLIPASE AS DETERMINED FROM 1H HOMONUCLEAR TWO-AND THREE-DIMENSIONAL NMR | | Descriptor: | HYDROXIDE ION, PORCINE PANCREATIC PROCOLIPASE B | | Authors: | Breg, J.N, Sarda, L, Cozzone, P.J, Rugani, N, Boelens, R, Kaptein, R. | | Deposit date: | 1994-06-08 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic procolipase as determined from 1H homonuclear two-dimensional and three-dimensional NMR.
Eur.J.Biochem., 227, 1995
|
|
6UNQ
 
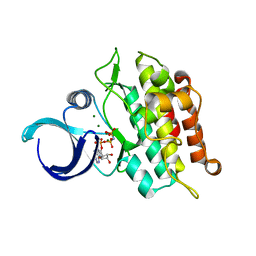 | | Kinase domain of ALK2-K493A with AMPPNP | | Descriptor: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNR
 
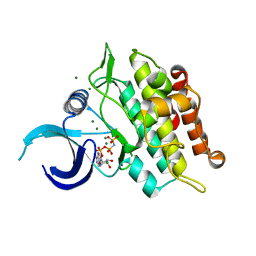 | | Kinase domain of ALK2-K492A/K493A with AMPPNP | | Descriptor: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNP
 
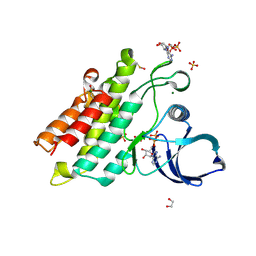 | | Crystal structure of the kinase domain of BMPR2-D485G | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bone morphogenetic protein receptor type-2, ... | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
1PIM
 
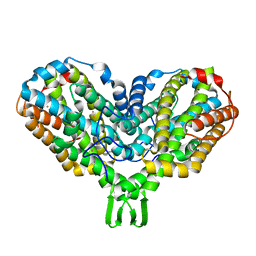 | | DITHIONITE REDUCED E. COLI RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT, D84E MUTANT | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Khidekel, N, Baldwin, J, Ley, B.A, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Ribonucleotide Reductase R2 Mutant that Accumulates a u-1,2-Peroxodiiron(III)
Intermediate during Oxygen Activation
J.Am.Chem.Soc., 122, 2000
|
|
6UNS
 
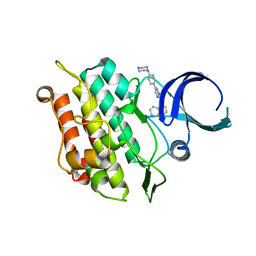 | | Kinase domain of ALK2-K492A/K493A with LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1 | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
1PE7
 
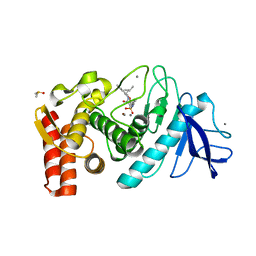 | | Thermolysin with bicyclic inhibitor | | Descriptor: | 2-(4-METHYLPHENOXY)ETHYLPHOSPHINATE, 3-METHYLBUTAN-1-AMINE, CALCIUM ION, ... | | Authors: | Juers, D, Yusuff, N, Bartlett, P.A, Matthews, B.W. | | Deposit date: | 2003-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conformational Constraint and Structural Complementarity in Thermolysin Inhibitors: Structures of Enzyme Complexes and Conclusions
To be Published
|
|
6TWG
 
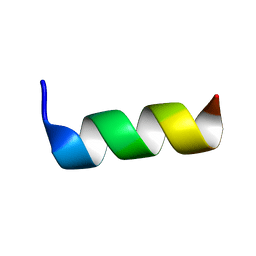 | | Solution structure of antimicrobial peptide, crabrolin Plus in the presence of Lipopolysaccharide | | Descriptor: | Crabrolin Plus, mutant of Crabrolin peptide | | Authors: | Cantini, F, Bouchemal, N, Savarin, P, Sette, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Effect of positive charges in the structural interaction of crabrolin isoforms with lipopolysaccharide.
J.Pept.Sci., 26, 2020
|
|
6UF2
 
 | |
