7CEH
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S/ mutant with the C-terminal three residues deletion in ligand ejecting form | | Descriptor: | Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Senga, A, Numoto, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-06-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Multiple structural states of Ca2+-regulated PET hydrolase, Cut190, and its correlation with activity and stability.
J.Biochem., 169, 2021
|
|
1PVZ
 
 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
1Q1U
 
 | | Crystal structure of human FHF1b (FGF12b) | | Descriptor: | SULFATE ION, fibroblast growth factor homologous factor 1 | | Authors: | Olsen, S.K, Garbi, M, Zampieri, N, Eliseenkova, A.V, Ornitz, D.M, Goldfarb, M, Mohammadi, M. | | Deposit date: | 2003-07-22 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs
J.Biol.Chem., 278, 2003
|
|
1Q67
 
 | | Crystal structure of Dcp1p | | Descriptor: | Decapping protein involved in mRNA degradation-Dcp1p | | Authors: | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | Deposit date: | 2003-08-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
1Q7X
 
 | | Solution structure of the alternatively spliced PDZ2 domain (PDZ2b) of PTP-Bas (hPTP1E) | | Descriptor: | PDZ2b domain of PTP-Bas (hPTP1E) | | Authors: | Kachel, N, Erdmann, K.S, Kremer, W, Wolff, P, Gronwald, W, Heumann, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-08-20 | | Release date: | 2003-12-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): Splicing induced modulation of ligand specificity.
J.Mol.Biol., 334, 2003
|
|
1QG7
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | STROMAL CELL-DERIVED FACTOR 1 ALPHA, SULFATE ION | | Authors: | Senda, T, Nandhagopal, N, Sugimoto, K, Mitsui, Y. | | Deposit date: | 1999-04-21 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of recombinant native SDF-1alpha with additional mutagenesis studies: an attempt at a more comprehensive interpretation of accumulated structure-activity relationship data.
J.Interferon Cytokine Res., 20, 2000
|
|
1QFZ
 
 | | PEA FNR Y308S MUTANT IN COMPLEX WITH NADPH | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), ... | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
1QKK
 
 | | Crystal structure of the receiver domain and linker region of DctD from Sinorhizobium meliloti | | Descriptor: | C4-DICARBOXYLATE TRANSPORT TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Meyer, M.G, Park, S, Zeringue, L, Staley, M, Mckinstry, M, Kaufman, R.I, Zhang, H, Yan, D, Yennawar, N, Farber, G.K, Nixon, B.T. | | Deposit date: | 1999-07-23 | | Release date: | 2000-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A dimeric two-component receiver domain inhibits the sigma54-dependent ATPase in DctD.
Faseb J., 15, 2001
|
|
1N26
 
 | | Crystal Structure of the extra-cellular domains of Human Interleukin-6 Receptor alpha chain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Varghese, J.N, Moritz, R.L, Lou, M.-Z, van Donkelaar, A, Ji, H, Ivancic, N, Branson, K.M, Hall, N.E, Simpson, R.J. | | Deposit date: | 2002-10-22 | | Release date: | 2002-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the extracellular domains of the human interleukin-6 receptor alpha-chain.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2GQV
 
 | |
1MVT
 
 | | Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',4',5'-TRIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate Reductase, SULFATE ION | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W.A, Gangjee, A. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of two polymorphic forms of a pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolate binary complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1M8L
 
 | |
1MN8
 
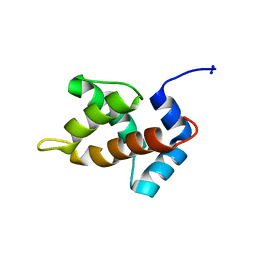 | | Structure of Moloney Murine Leukaemia Virus Matrix Protein | | Descriptor: | Core protein p15 | | Authors: | Riffel, N, Harlos, K, Iourin, O, Rao, Z, Kingsman, A, Stuart, D, Fry, E. | | Deposit date: | 2002-09-05 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of Moloney murine leukaemia virus matrix protein and its relationship to other retroviral matrix proteins.
Structure, 10, 2002
|
|
1M4X
 
 | | PBCV-1 virus capsid, quasi-atomic model | | Descriptor: | PBCV-1 virus capsid | | Authors: | Nandhagopal, N, Simpson, A.A, Gurnon, J.R, Yan, X, Baker, T.S, Graves, M.V, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2002-07-05 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | The Structure and Evolution of the Major Capsid Protein of a Large,
Lipid containing, DNA virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MDF
 
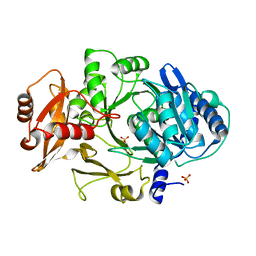 | | CRYSTAL STRUCTURE OF DhbE IN ABSENCE OF SUBSTRATE | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase, SULFATE ION | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MFU
 
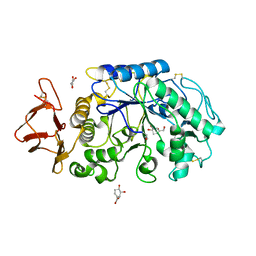 | | Probing the role of a mobile loop in human salivary amylase: Structural studies on the loop-deleted mutant | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | Authors: | Ramasubbu, N, Ragunath, C, Mishra, P.J. | | Deposit date: | 2002-08-13 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of a mobile loop in substrate binding and enzyme activity of
human salivary amylase.
J.Mol.Biol., 325, 2003
|
|
1QTK
 
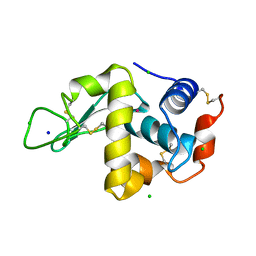 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF KRYPTON (55 BAR) | | Descriptor: | CHLORIDE ION, KRYPTON, LYSOZYME, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-06-28 | | Release date: | 1999-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1QKH
 
 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S19 FROM THERMUS THERMOPHILUS | | Descriptor: | 30S RIBOSOMAL PROTEIN S19 | | Authors: | Helgstrand, M, Rak, A.V, Allard, P, Davydova, N, Garber, M.B, Hard, T. | | Deposit date: | 1999-07-20 | | Release date: | 1999-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosomal protein S19 from Thermus thermophilus.
J. Mol. Biol., 292, 1999
|
|
1Q8I
 
 | | Crystal structure of ESCHERICHIA coli DNA Polymerase II | | Descriptor: | DNA polymerase II | | Authors: | Brunzelle, J.S, Muchmore, C.R.A, Mashhoon, N, Blair-Johnson, M, Shuvalova, L, Goodman, M.F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Escherichia Coli DNA Polymerase II
To be Published
|
|
1QSH
 
 | | MAGNESIUM(II)-AND ZINC(II)-PROTOPORPHYRIN IX'S STABILIZE THE LOWEST OXYGEN AFFINITY STATE OF HUMAN HEMOGLOBIN EVEN MORE STRONGLY THAN DEOXYHEME | | Descriptor: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miyazaki, G, Morimoto, H, Yun, K.-M, Park, S.-Y, Nakagawa, A, Minagawa, H, Shibayama, N. | | Deposit date: | 1999-06-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Magnesium(II) and zinc(II)-protoporphyrin IX's stabilize the lowest oxygen affinity state of human hemoglobin even more strongly than deoxyheme.
J.Mol.Biol., 292, 1999
|
|
1QTR
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE PROLYL AMINOPEPTIDASE FROM SERRATIA MARCESCENS | | Descriptor: | PROLYL AMINOPEPTIDASE | | Authors: | Yoshimoto, T, Kabashima, T, Uchikawa, K, Inoue, T, Tanaka, N. | | Deposit date: | 1999-06-28 | | Release date: | 1999-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of prolyl aminopeptidase from Serratia marcescens.
J.Biochem.(Tokyo), 126, 1999
|
|
1QQU
 
 | |
2J9X
 
 | | Tryptophan Synthase in complex with GP, alpha-D,L-glycerol-phosphate, Cs, pH6.5 - alpha aminoacrylate form - (GP)E(A-A) | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ngo, H, Kimmich, N, Harris, R, Niks, D, Blumenstein, L, Kulik, V, Barends, T.R, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-11-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Regulation of Substrate Channeling in Tryptophan Synthase: Modulation of the L-Serine Reaction in Stage I of the Beta-Reaction by Alpha-Site Ligands.
Biochemistry, 46, 2007
|
|
2K8U
 
 | | Solution NMR structure of trans-4-hydroxynonenal derived dG adduct of (6S,8R,11S)-configuration matched with dC | | Descriptor: | (2S,5R)-5-pentyltetrahydrofuran-2-ol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DCP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Qi, N, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2'-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5'-CpG-3' sequence.
Biochemistry, 47, 2008
|
|
234L
 
 | | T4 LYSOZYME MUTANT M106L | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-07 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
