9AYK
 
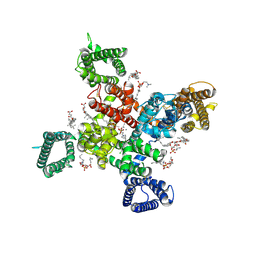 | | Cryo-EM structure of human Cav3.2 with ML218 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5-dichloro-N-{[(1R,5S,6r)-3-(3,3-dimethylbutyl)-3-azabicyclo[3.1.0]hexan-6-yl]methyl}benzamide, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYG
 
 | | Cryo-EM structure of apo state human Cav3.2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYH
 
 | | Cryo-EM structure of human Cav3.2 with TTA-A2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(4-cyclopropylphenyl)-N-{(1R)-1-[5-(2,2,2-trifluoroethoxy)pyridin-2-yl]ethyl}acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYL
 
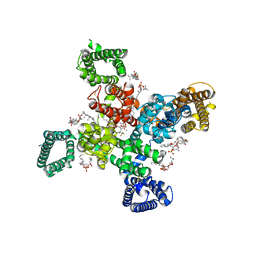 | | Cryo-EM structure of human Cav3.2 with ACT-709478 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
8FBZ
 
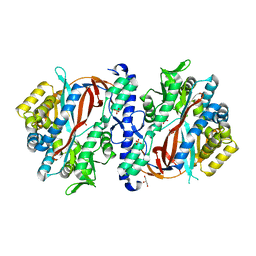 | | Crystal Structure of apo human Glutathione Synthetase Y270E | | Descriptor: | GLYCEROL, Glutathione synthetase, SULFATE ION | | Authors: | Stanford, S.M, Santelli, E, Sankaran, B, Murali, R, Bottini, N. | | Deposit date: | 2022-11-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting prostate tumor low-molecular weight tyrosine phosphatase for oxidation-sensitizing therapy.
Sci Adv, 10, 2024
|
|
1L4D
 
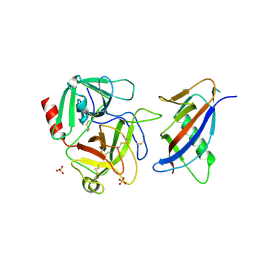 | | CRYSTAL STRUCTURE OF MICROPLASMINOGEN-STREPTOKINASE ALPHA DOMAIN COMPLEX | | Descriptor: | PLASMINOGEN, STREPTOKINASE, SULFATE ION | | Authors: | Wakeham, N, Terzyan, S, Zhai, P, Loy, J.A, Tang, J, Zhang, X.C. | | Deposit date: | 2002-03-04 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of deletion of streptokinase residues 48-59 on plasminogen activation
PROTEIN ENG., 15, 2002
|
|
9BBM
 
 | |
1LBB
 
 | | Crystal structure of the GluR2 ligand binding domain mutant (S1S2J-N754D) in complex with kainate at 2.1 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamine receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LOL
 
 | | Crystal structure of orotidine monophosphate decarboxylase complex with XMP | | Descriptor: | 1,3-BUTANEDIOL, XANTHOSINE-5'-MONOPHOSPHATE, orotidine 5'-monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
8WE6
 
 | | Human L-type voltage-gated calcium channel Cav1.2 at 2.9 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
1LBC
 
 | | Crystal structure of GluR2 ligand binding core (S1S2J-N775S) in complex with cyclothiazide (CTZ) as well as glutamate at 1.8 A resolution | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamine Receptor 2, ... | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LF0
 
 | | Crystal Structure of RasA59G in the GTP-bound form | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hall, B.E, Bar-Sagi, D, Nassar, N. | | Deposit date: | 2002-04-10 | | Release date: | 2002-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis for the Transition from Ras-GTP to Ras-GDP
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8WEA
 
 | | Human L-type voltage-gated calcium channel Cav1.2 (Class II) in the presence of pinaverium at 3.2 Angstrom resolution | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE9
 
 | | Human L-type voltage-gated calcium channel Cav1.2 (Class I) in the presence of pinaverium at 3.0 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE8
 
 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine, amlodipine and pinaverium at 2.9 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
1LJX
 
 | | THE STRUCTURE OF D(TPGPCPGPCPA)2 AT 293K: COMPARISON OF THE EFFECT OF SEQUENCE AND TEMPERATURE | | Descriptor: | 5'-D(*TP*GP*CP*GP*CP*A)-3', MAGNESIUM ION | | Authors: | Thiyagarajan, S, Satheesh Kumar, P, Rajan, S.S, Gautham, N. | | Deposit date: | 2002-04-23 | | Release date: | 2002-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of d(TGCGCA)2 at 293 K: comparison of the effects of sequence and temperature.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KZL
 
 | | Riboflavin Synthase from S.pombe bound to Carboxyethyllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, MERCURY (II) ION, Riboflavin Synthase | | Authors: | Gerhardt, S, Schott, A.K, Kairies, N, Cushman, M, Illarionov, B, Eisenreich, W, Bacher, A, Huber, R, Steinbacher, S, Fischer, M. | | Deposit date: | 2002-02-07 | | Release date: | 2002-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on the Reaction Mechanism of Riboflavin Synthase; X-Ray Crystal Structure of a Complex with 6-Carboxyethyl-7-Oxo-8-Ribityllumazine
STRUCTURE, 10, 2002
|
|
8VTU
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-66, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | Descriptor: | 1-cyclopropyl-7-[(4-{[3-({(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-14-[({3-[5-(methylcarbamoyl)pyridin-3-yl]prop-2-yn-1-yl}oxy)imino]-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl}oxy)-3-oxopropyl]amino}butyl)amino]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
8VTY
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with ciprofloxacin and protein Y at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 16S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
8VTW
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-128 and protein Y at 2.35A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-cyclopropyl-7-(4-{4-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]butyl}piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
1L3A
 
 | |
8W96
 
 | | SmChiA with diacetyl chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A, GLYCEROL | | Authors: | Yoshiko, T, Akihiko, N. | | Deposit date: | 2023-09-04 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SmChiA with diacetyl chitobiose
To Be Published
|
|
1L6G
 
 | | Alanine racemase bound with N-(5'-phosphopyridoxyl)-D-alanine | | Descriptor: | N-(5'-PHOSPHOPYRIDOXYL)-D-ALANINE, alanine racemase | | Authors: | Watanabe, A, Yoshimura, T, Mikami, B, Hayashi, H, Kagamiyama, H, Esaki, N. | | Deposit date: | 2002-03-10 | | Release date: | 2002-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5'-phosphopyridoxyl)alanine.
J.Biol.Chem., 277, 2002
|
|
9BBL
 
 | |
6YCD
 
 | | Structure the ananain protease from Ananas comosus covalently bound to the TLCK inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
