2PB5
 
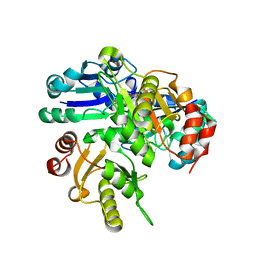 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | FORMIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamamoto, H, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2PB6
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | FORMIC ACID, GLYCEROL, Probable diphthine synthase, ... | | Authors: | Yamamoto, H, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
7BQE
 
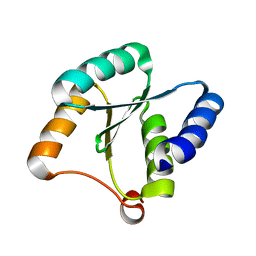 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | Descriptor: | NF3 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
6LZ0
 
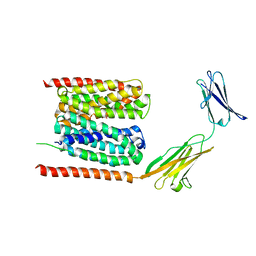 | | Cryo-EM structure of human MCT1 in complex with Basigin-2 in the presence of lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
6MEA
 
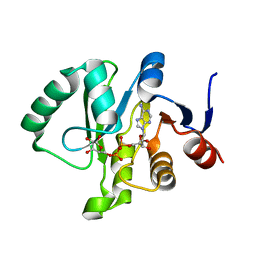 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate ribose (ADP-ribose) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6LL6
 
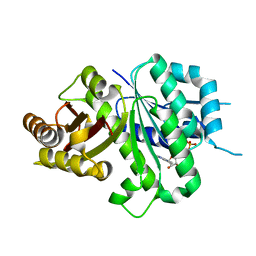 | | Crsyal structure of EcFtsZ (residues 11-316) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yoshizawa, T, Fujita, J, Terakada, H, Ozawa, M, Kuroda, N, Tanaka, S, Uehara, R, Matsumura, H. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the cell-division protein FtsZ from Klebsiella pneumoniae and Escherichia coli.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3KLS
 
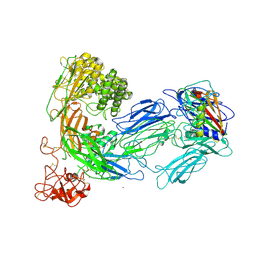 | | Structure of complement C5 in complex with SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KM9
 
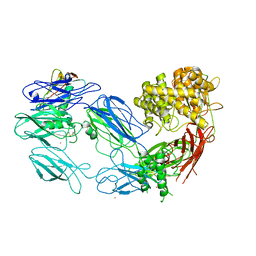 | | Structure of complement C5 in complex with the C-terminal beta-grasp domain of SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6M9Y
 
 | |
6MAS
 
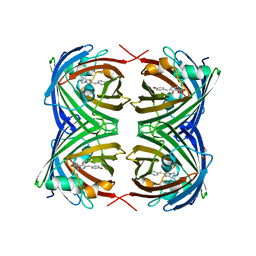 | | X-ray Structure of Branchiostoma floridae fluorescent protein lanFP10G | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V, Pletnev, S. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Factors Enabling Successful GFP-Like Proteins with Alanine as the Third Chromophore-Forming Residue.
J. Mol. Biol., 431, 2019
|
|
6ME9
 
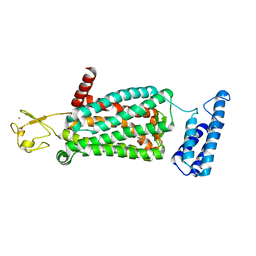 | | XFEL crystal structure of human melatonin receptor MT2 in complex with ramelteon | | Descriptor: | N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
6M0V
 
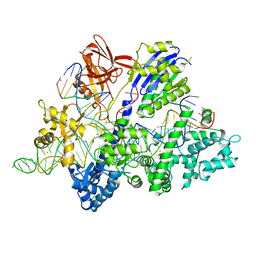 | | Crsytal structure of streptococcus thermophilus Cas9 in complex with the GGAA PAM | | Descriptor: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-22 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
1WQS
 
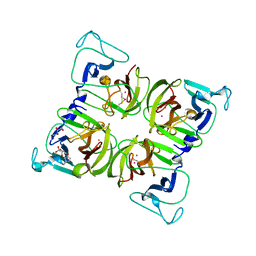 | | Crystal structure of Norovirus 3C-like protease | | Descriptor: | 3C-like protease, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Nakamura, K, Someya, Y, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-10-01 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A norovirus protease structure provides insights into active and substrate binding site integrity
J.Virol., 79, 2005
|
|
6LL5
 
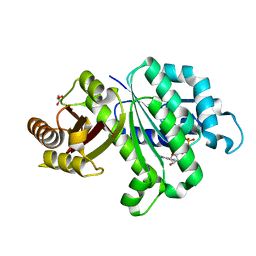 | | Crystal structure of KpFtsZ (residues 11-316) | | Descriptor: | Cell division protein FtsZ, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yoshizawa, T, Fujita, J, Terakado, H, Ozawa, M, Kuroda, N, Tanaka, S, Uehara, R, Matsumura, H. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the cell-division protein FtsZ from Klebsiella pneumoniae and Escherichia coli.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6M5V
 
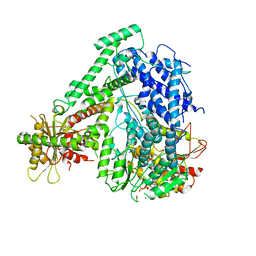 | | The coordinate of the hexameric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6LQW
 
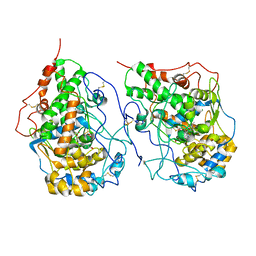 | | Crystal structure of a dimeric yak lactoperoxidase at 2.59 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Viswanathan, V, Pandey, S.N, Ahmad, N, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a dimeric yak lactoperoxidase at 2.59 A resolution.
To Be Published
|
|
6MD5
 
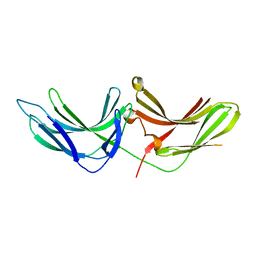 | |
6ME7
 
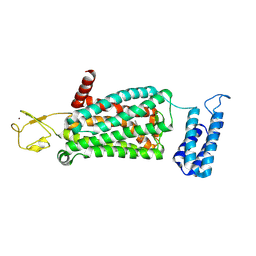 | | XFEL crystal structure of human melatonin receptor MT2 (H208A) in complex with 2-phenylmelatonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ... | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
1WLK
 
 | |
6M5E
 
 | | Human serum albumin with cyclic peptide dalbavancin | | Descriptor: | (2S,5S)-5-azanyl-3,4,6-tris(oxidanyl)oxane-2-carboxylic acid, 10-METHYLUNDECANOIC ACID, 2-amino-2-deoxy-beta-D-altropyranuronic acid, ... | | Authors: | Ito, S, Senoo, A, Nagatoishi, S, Ohue, M, Yamamoto, M, Tsumoto, K, Wakui, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Mechanism of Human Serum Albumin Complexed with Cyclic Peptide Dalbavancin.
J.Med.Chem., 63, 2020
|
|
6M9X
 
 | |
6MEC
 
 | | Structure of a group II intron retroelement after DNA integration | | Descriptor: | MAGNESIUM ION, Maturase reverse transcriptase, SODIUM ION, ... | | Authors: | Haack, D, Yan, X, Zhang, C, Hingey, J, Lyumkis, D, Baker, T.S, Toor, N. | | Deposit date: | 2018-09-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures of a Group II Intron Reverse Splicing into DNA.
Cell, 178, 2019
|
|
6ME6
 
 | | XFEL crystal structure of human melatonin receptor MT2 in complex with 2-phenylmelatonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ... | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
6MEL
 
 | | Succinyl-CoA synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, CITRIC ACID, Succinate--CoA ligase [ADP-forming] subunit beta, ... | | Authors: | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Succinyl-CoA synthase from Campylobacter jejuni
to be published
|
|
6ME8
 
 | | XFEL crystal structure of human melatonin receptor MT2 (N86D) in complex with 2-phenylmelatonin | | Descriptor: | N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
