5ERJ
 
 | |
6VYP
 
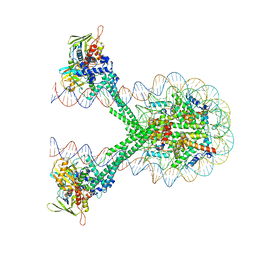 | | Crystal structure of the LSD1/CoREST histone demethylase bound to its nucleosome substrate | | Descriptor: | DNA (191-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A type 1, ... | | Authors: | Kim, S, Zhu, J, Eek, P, Yennawar, N, Song, T. | | Deposit date: | 2020-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.99 Å) | | Cite: | Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate.
Mol.Cell, 78, 2020
|
|
5EK9
 
 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline inhibitor | | Descriptor: | Bromodomain-containing protein 2, propan-2-yl ~{N}-[(2~{S},4~{R})-1-ethanoyl-6-(furan-2-yl)-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Picaud, S, Fedorov, O, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploiting a water network to achieve enthalpy-driven, bromodomain-selective BET inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
5EKQ
 
 | | The structure of the BamACDE subcomplex from E. coli | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, Outer membrane protein assembly factor BamD, ... | | Authors: | Bakelar, J, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.392 Å) | | Cite: | The structure of the beta-barrel assembly machinery complex.
Science, 351, 2016
|
|
6DQV
 
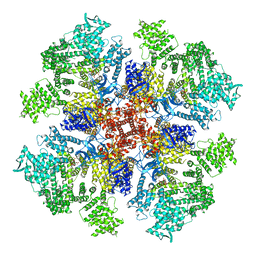 | | Class 2 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1ANS
 
 | |
6DRC
 
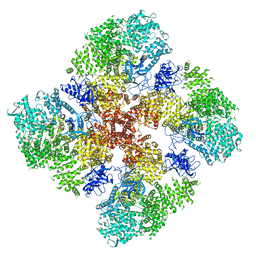 | | High IP3 Ca2+ human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6E14
 
 | | Handover mechanism of the growing pilus by the bacterial outer membrane usher FimD | | Descriptor: | Chaperone protein FimC, Fimbrial biogenesis outer membrane usher protein, Protein FimF, ... | | Authors: | Du, M, Yuan, Z, Yu, H, Henderson, N, Sarowar, S, Zhao, G, Werneburg, G.T, Thanassi, D.G, Li, H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD.
Nature, 562, 2018
|
|
6VP0
 
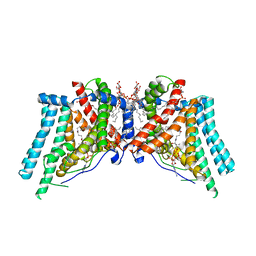 | | Human Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Diacylglycerol O-acyltransferase 1, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Qian, H, Han, Y, Nian, Y, Ren, Z, Zhang, H, Hu, L, Prasad, B.V.V, Yan, N, Zhou, M. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-13 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of human diacylglycerol O-acyltransferase 1.
Nature, 581, 2020
|
|
6VRB
 
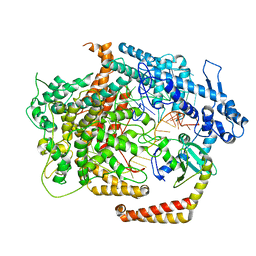 | | Cryo-EM structure of AcrVIA1-Cas13(crRNA) complex | | Descriptor: | AcrVIA1, CRISPR-associated endoribonuclease Cas13a, RNA (52-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
6WCB
 
 | | Human open state TMEM175 in CsCl | | Descriptor: | CESIUM ION, Endosomal/lysosomal potassium channel TMEM175 | | Authors: | Oh, S, Paknejad, N, Hite, R.K. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Gating and selectivity mechanisms for the lysosomal K + channel TMEM175.
Elife, 9, 2020
|
|
6WYC
 
 | |
1AGQ
 
 | | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR FROM RAT | | Descriptor: | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR | | Authors: | Eigenbrot, C, Gerber, N. | | Deposit date: | 1997-03-25 | | Release date: | 1997-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of glial cell-derived neurotrophic factor at 1.9 A resolution and implications for receptor binding.
Nat.Struct.Biol., 4, 1997
|
|
5ER3
 
 | | Crystal structure of ABC transporter system solute-binding protein from Rhodopirellula baltica SH 1 | | Descriptor: | CALCIUM ION, GLYCEROL, Sugar ABC transporter, ... | | Authors: | Chang, C, Duke, N, Endres, M, Mack, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Crystal structure of ABC transporter system solute-binding protein from Rhodopirellula baltica SH 1
To Be Published
|
|
6WCC
 
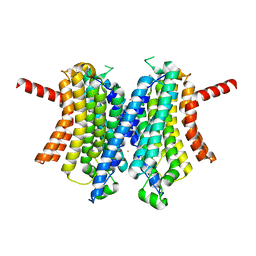 | | Human closed state TMEM175 in CsCl | | Descriptor: | CESIUM ION, Endosomal/lysosomal potassium channel TMEM175 | | Authors: | Oh, S, Paknejad, N, Hite, R.K. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Gating and selectivity mechanisms for the lysosomal K + channel TMEM175.
Elife, 9, 2020
|
|
6WHT
 
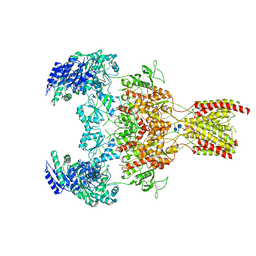 | | GluN1b-GluN2B NMDA receptor in active conformation at 4.4 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WIK
 
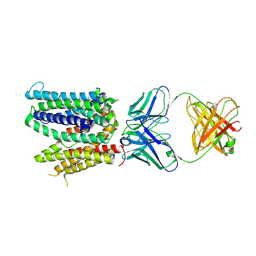 | | Cryo-EM structure of SLC40/ferroportin with Fab in the presence of hepcidin | | Descriptor: | 11F9 Fab heavy-chain, 11F9 Fab light-chain, Solute carrier family 40 protein | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
8B56
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GD-9 | | Descriptor: | (2~{S})-4-(2-chloranylethanoyl)-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5, BROMIDE ION, ... | | Authors: | Straeter, N, Muller, C.E, Claff, T, Sylvester, K, Weisse, R, Gao, S, Song, L, Liu, X, Zhan, P. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives as Covalent SARS-CoV-2 Main Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
5EX7
 
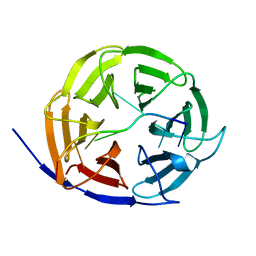 | | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA | | Descriptor: | Brain tumor protein, RNA (5'-R(P*UP*UP*UP*GP*UP*UP*GP*U)-3') | | Authors: | Wang, Y, Yu, Z, Wang, M, Liu, C.P, Yang, N, Xu, R.M. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA
To Be Published
|
|
6WOQ
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody HC1AM and non neutralizing antibody E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.667 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
1RUY
 
 | | 1930 Swine H1 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Skehel, J.J, Gamblin, S.J, Haire, L.F, Russell, R.J, Stevens, D.J, Xiao, B, Ha, Y, Vasisht, N, Steinhauer, D.A, Daniels, R.S. | | Deposit date: | 2003-12-12 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure and receptor binding properties of the 1918 influenza hemagglutinin.
Science, 303, 2004
|
|
1ROW
 
 | | Structure of SSP-19, an MSP-domain protein like family member in Caenorhabditis elegans | | Descriptor: | MSP-domain protein like family member | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, J, Lu, S, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sperm-specific protein SSP-19 from Caenorhabditis elegans.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6WPF
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and d4T | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA Primer 21-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Post-Catalytic Complexes with Emtricitabine or Stavudine and HIV-1 Reverse Transcriptase Reveal New Mechanistic Insights for Nucleotide Incorporation and Drug Resistance.
Molecules, 25, 2020
|
|
5F5U
 
 | | Crystal structure of the Snu23-Prp38-MFAP1(217-258) complex of Chaetomium thermophilum | | Descriptor: | Prp38, Putative uncharacterized protein, Zinc finger domain-containing protein | | Authors: | Ulrich, A.K.C, Seeger, M, Bartlick, N, Wahl, M.C. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | Scaffolding in the Spliceosome via Single alpha Helices.
Structure, 24, 2016
|
|
1RVZ
 
 | | 1934 H1 Hemagglutinin in complex with LSTC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Gamblin, S.J, Haire, L.F, Russell, R.J, Stevens, D.J, Xiao, B, Ha, Y, Vasisht, N, Steinhauer, D.A, Daniels, R.S, Elliot, A, Wiley, D.C, Skehel, J.J. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure and receptor binding properties of the 1918 influenza hemagglutinin.
Science, 303, 2004
|
|
