2KVP
 
 | | Solution Structure of the R10 Domain of Talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Gingras, A.R, Bate, N, Critchley, D.R, Barsukov, I.L, Roberts, G.C. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The domain structure of talin: Residues 1815-1973 form a five-helix bundle containing a cryptic vinculin-binding site.
Febs Lett., 584, 2010
|
|
2KXN
 
 | | NMR structure of human Tra2beta1 RRM in complex with AAGAAC RNA | | Descriptor: | 5'-R(*AP*AP*GP*AP*AP*C)-3', Transformer-2 protein homolog beta | | Authors: | Clery, A, Jayne, S, Benderska, N, Dominguez, C, Stamm, S, Allain, F.H.-T. | | Deposit date: | 2010-05-10 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1
Nat.Struct.Mol.Biol., 18, 2011
|
|
2KSH
 
 | |
2KYL
 
 | | Solution structure of MAST2-PDZ complexed with the C-terminus of PTEN | | Descriptor: | C-terminus of Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, Microtubule-associated serine/threonine-protein kinase 2 | | Authors: | Terrien, E, Wolff, N, Cordier, F, Simenel, C, Lafon, M, Delepierre, M. | | Deposit date: | 2010-06-01 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MAST2-PDZ complexed with the C-terminus of PTEN
To be Published
|
|
2K90
 
 | | Dimeric solution structure of the DNA loop d(TGCTTCGT) | | Descriptor: | 5'-D(*TP*GP*CP*TP*TP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-29 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
6QN9
 
 | | Structure of bovine anti-RSV Fab B4 | | Descriptor: | GLYCEROL, Heavy chain, SULFATE ION, ... | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
2KNE
 
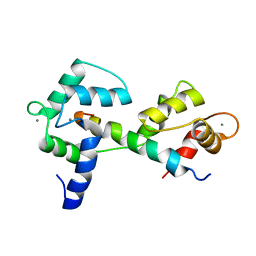 | | Calmodulin wraps around its binding domain in the plasma membrane CA2+ pump anchored by a novel 18-1 motif | | Descriptor: | ATPase, Ca++ transporting, plasma membrane 4, ... | | Authors: | Juranic, N, Atanasova, E, Filoteo, A.G, Macura, S, Prendergast, F.G, Penniston, J.T, Strehler, E.E. | | Deposit date: | 2009-08-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Calmodulin wraps around its binding domain in the plasma membrane Ca2+ pump anchored by a novel 18-1 motif.
J.Biol.Chem., 285, 2010
|
|
7MSK
 
 | | ThuS glycosin S-glycosyltransferase | | Descriptor: | Glyco_trans_2-like domain-containing protein, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
7MSN
 
 | | SunS glycosin S-glycosyltransferase | | Descriptor: | SPbeta prophage-derived glycosyltransferase SunS, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
7MSP
 
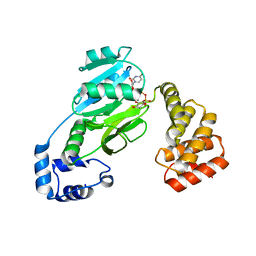 | | SunS glycosin S-glycosyltransferase | | Descriptor: | MAGNESIUM ION, SPbeta prophage-derived glycosyltransferase SunS, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
4OIM
 
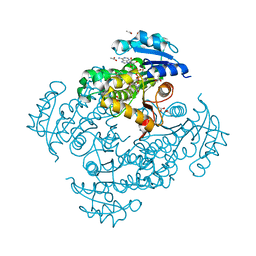 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT119 in 2.4 M acetate | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-Dependent Diaryl Ether Inhibitors of InhA: Structure-Activity Relationship Studies of Enzyme Inhibition, Antibacterial Activity, and in vivo Efficacy.
Chemmedchem, 9, 2014
|
|
7MHE
 
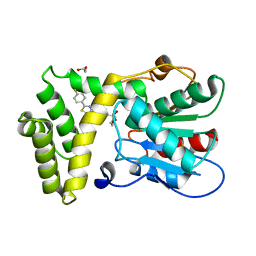 | | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7957 | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[2-(4-fluorophenyl)-1,3-thiazol-4-yl]benzene-1-sulfonyl}morpholine, Fatty acid synthase | | Authors: | Aleshin, A.E, Lambert, L, Liddington, R.C, Cosford, N. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7635
To Be Published
|
|
4OMT
 
 | |
7MHD
 
 | | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7635 | | Descriptor: | Fatty acid synthase, N,N-diethyl-4-{2-[(2-fluorophenyl)methyl]-1,3-thiazol-4-yl}benzene-1-sulfonamide | | Authors: | Aleshin, A.E, Lambert, L, Liddington, R.C, Cosford, N. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7635
To Be Published
|
|
7MLK
 
 | | Crystal structure of human PI3Ka (p110a subunit) with MMV085400 bound to the active site determined at 2.9 angstroms resolution | | Descriptor: | 4-[6-(3,4,5-trimethoxyanilino)pyrazin-2-yl]benzamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Krake, S.H, Martinez, P.D.G, Poggi, M.L, Ferreira, M.S, Aguiar, A.C.C, Souza, G.E, Wenlock, M, Jones, B, Steinbrecher, T, Day, T, McPhail, J, Burke, J, Yeo, T, Mok, S, Uhlemann, A.C, Fidock, D.A, Chen, P, Grodsky, N, Deng, Y.L, Guido, R.V.C, Campbell, S.F, Willis, P.A, Dias, L.C. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazines as potent PI4K inhibitors with antimalarial activity
To Be Published
|
|
6QZ8
 
 | | Structure of Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
2K97
 
 | | Dimeric solution structure of the cyclic octamer d(pCGCTCCGT) | | Descriptor: | 5'-D(P*CP*GP*CP*TP*CP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-30 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
7MCO
 
 | | Crystal Structure of Tetur04g02350 | | Descriptor: | UDP-glycosyltransferase 203A2, URIDINE-5'-DIPHOSPHATE | | Authors: | Danehsian, L, Kluza, A, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Tetur04g02350
To Be Published
|
|
6QU9
 
 | | Fab fragment of an antibody that inhibits polymerisation of alpha-1-antitrypsin | | Descriptor: | FAB 4B12 heavy chain, FAB 4B12 light chain, GLYCEROL, ... | | Authors: | Jagger, A.M, Heyer-Chauhan, N, Lomas, D.A, Irving, J.A. | | Deposit date: | 2019-02-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for Z alpha 1 -antitrypsin polymerization in the liver.
Sci Adv, 6, 2020
|
|
2LBD
 
 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO ALL-TRANS RETINOIC ACID | | Descriptor: | RETINOIC ACID, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Renaud, J.-P, Rochel, N, Ruff, M, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1997-08-19 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid.
Nature, 378, 1995
|
|
6QYP
 
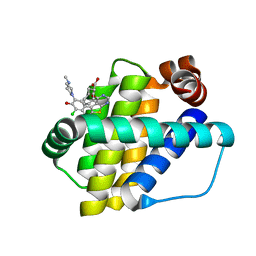 | | Structure of Mcl-1 in complex with compound 13 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
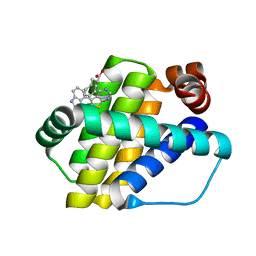 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
2L84
 
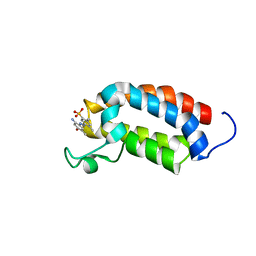 | | Solution NMR structures of CBP bromodomain with small molecule j28 | | Descriptor: | 5-[(E)-(2-amino-4-hydroxy-5-methylphenyl)diazenyl]-2,4-dimethylbenzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
6QYK
 
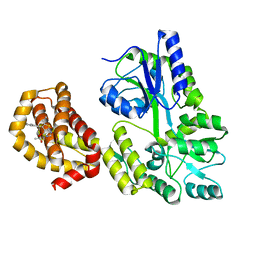 | | Structure of MBP-Mcl-1 in complex with compound 7a | | Descriptor: | (2~{R})-2-[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]oxypropanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
2L3H
 
 | | NMR Structure in a Membrane Environment Reveals Putative Amyloidogenic Regions of the SEVI Precursor Peptide PAP248-286 | | Descriptor: | Prostatic acid phosphatase | | Authors: | Ramamoorthy, A, Nanga, R, Brender, J, Vivekanandan, S, Popovych, N. | | Deposit date: | 2010-09-13 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure in a membrane environment reveals putative amyloidogenic regions of the SEVI precursor peptide PAP(248-286).
J.Am.Chem.Soc., 131, 2009
|
|
