7NHE
 
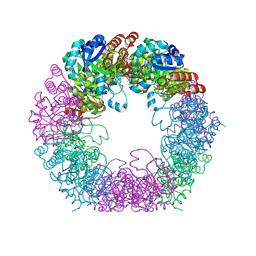 | | Crystal structure of Arabidopsis thaliana Pdx1K166R-I333 complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, [(~{E},4~{S})-4-azanyl-3-oxidanylidene-pent-1-enyl] dihydrogen phosphate | | Authors: | Rodrigues, M.J, Zhang, Y, Bolton, R, Evans, G, Giri, N, Royant, A, Begley, T, Ealick, S.E, Tews, I. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Trapping and structural characterisation of a covalent intermediate in vitamin B 6 biosynthesis catalysed by the Pdx1 PLP synthase.
Rsc Chem Biol, 3, 2022
|
|
2KZK
 
 | | Solution structure of alpha-mannosidase binding domain of Atg34 | | Descriptor: | Uncharacterized protein YOL083W | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
6ZXP
 
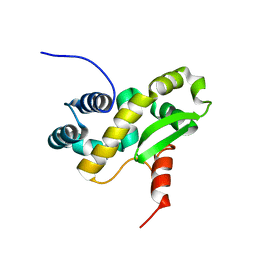 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20 fused to a short peptide from the viral DNA polymerase E9. | | Descriptor: | DNA polymerase processivity factor component A20,DNA polymerase processivity factor component E9 | | Authors: | Bersch, B, Tarbouriech, N, Burmeister, W, Iseni, F. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
6ZYC
 
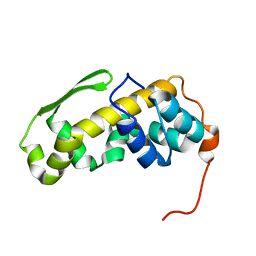 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20. | | Descriptor: | DNA polymerase processivity factor component A20 | | Authors: | Bersch, B, Iseni, F, Burmeister, W, Tarbouriech, N. | | Deposit date: | 2020-07-31 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
2LDC
 
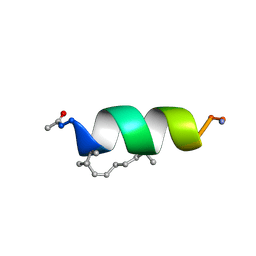 | | Solution structure of the estrogen receptor-binding stapled peptide SP1 (Ac-HXILHXLLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP1 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
7NLV
 
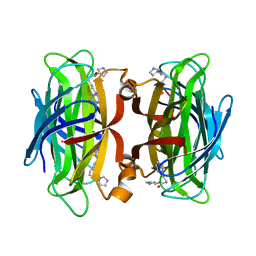 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE II | | Descriptor: | 5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-N-((S)-pyrrolidin-3-yl)pentanamide, Streptavidin | | Authors: | Nodling, A.R, Santi, N, Tsai, Y.H, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The role of streptavidin and its variants in catalysis by biotinylated secondary amines.
Org.Biomol.Chem., 19, 2021
|
|
7NBP
 
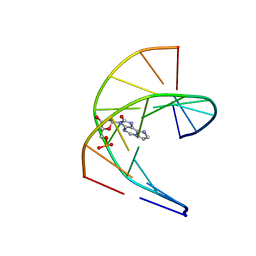 | | Solution structure of DNA duplex containing a 7,8-dihydro-8-oxo-1,N6-ethenoadenine base modification that induces exclusively A->T transversions in Escherichia coli | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*(EDA)P*CP*AP*TP*G)-3') | | Authors: | Aralov, A.V, Gubina, N, Cabrero, C, Tsvetkov, V.B, Turaev, A.V, Fedeles, B.I, Croy, R.G, Isaakova, E.A, Melnik, D, Dukova, S, Khrulev, A.A, Varizhuk, A.M, Gonzalez, C, Zatsepin, T.S, Essigmann, J.M. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 7,8-Dihydro-8-oxo-1,N6-ethenoadenine: an exclusively Hoogsteen-paired thymine mimic in DNA that induces A→T transversions in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
5EDK
 
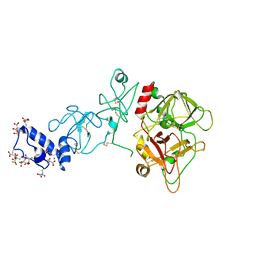 | | Crystal structure of prothrombin deletion mutant residues 146-167 ( Form II ). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | How the Linker Connecting the Two Kringles Influences Activation and Conformational Plasticity of Prothrombin.
J.Biol.Chem., 291, 2016
|
|
2M0G
 
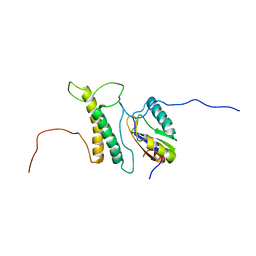 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2MHZ
 
 | | Structure of Exocyclic S,S N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(SDE)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
8A7N
 
 | | Crystal Structure of human Brachyury G177D variant in complex with (S)-N-(3-aminopropyl)-3-((1-(2-fluorophenyl)-2-oxopyrrolidin-3-yl)amino)-N-methylbenzamide (CF-2-125) | | Descriptor: | N-(3-azanylpropyl)-3-[[(3S)-1-(2-fluorophenyl)-2-oxidanylidene-pyrrolidin-3-yl]amino]-N-methyl-benzamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-06-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with (S)-N-(3-aminopropyl)-3-((1-(2-fluorophenyl)-2-oxopyrrolidin-3-yl)amino)-N-methylbenzamide (CF-2-125)
To Be Published
|
|
7N9J
 
 | | Crystal structure of H2DB in complex with HSF2 melanoma neoantigen | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the complex between H2DB and melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
7NA5
 
 | | Structure of the H2DB-TCR ternary complex with HSF2 melanoma neoantigen | | Descriptor: | 47BE7 TCR alpha chain, 47BE7 TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the TCR-H2DB ternary complex with melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
3VHU
 
 | |
2LZ6
 
 | | Distinct ubiquitin binding modes exhibited by sh3 domains: molecular determinants and functional implications | | Descriptor: | CD2-associated protein, Ubiquitin | | Authors: | Ortega-Roldan, J, Azuaga, A, Blackledge, M, Van Nuland, N. | | Deposit date: | 2012-09-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct Ubiquitin Binding Modes Exhibited by SH3 Domains: Molecular Determinants and Functional Implications.
Plos One, 8, 2013
|
|
2LF1
 
 | | Solution structure of L. casei dihydrofolate reductase complexed with NADPH, 30 structures | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Polshakov, V, Feeney, J, Birdsall, B, Kovalevskaya, N. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structures of apo L. casei dihydrofolate reductase and its complexes with trimethoprim and NADPH: contributions to positive cooperative binding from ligand-induced refolding, conformational changes, and interligand hydrophobic interactions
Biochemistry, 50, 2011
|
|
2MHX
 
 | | Structure of Exocyclic R,R N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(RBD)P*AP*GP*AP*AP*G)-3'), 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2MNX
 
 | | Major groove orientation of the (2S)-N6-(2-hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA adduct induced by 1,2-epoxy-3-butene | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(6HB)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Kotapati, S, Turo, M, Tretyakova, N, Stone, M.P. | | Deposit date: | 2014-04-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major Groove Orientation of the (2S)-N(6)-(2-Hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA Adduct Induced by 1,2-Epoxy-3-butene.
Chem.Res.Toxicol., 27, 2014
|
|
7NMO
 
 | | Crystal structure of beta-2-microglobulin D76A mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMR
 
 | | Crystal structure of beta-2-microglobulin D76S mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMT
 
 | | Crystal structure of beta-2-microglobulin D76G mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMC
 
 | | Crystal structure of beta-2-microglobulin D76E mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL, TRIETHYLENE GLYCOL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMV
 
 | | Crystal structure of beta-2-microglobulin D76Q mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMY
 
 | | Crystal structure of beta-2-microglobulin D76Y mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NN5
 
 | | Crystal structure of beta-2-microglobulin D76K mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-24 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
