2QHW
 
 | | Crystal structure of a complex of phospholipase A2 with a gramine derivative at 2.2 resolution | | Descriptor: | 3-{3-[(DIMETHYLAMINO)METHYL]-1H-INDOL-7-YL}PROPAN-1-OL, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-07-03 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of a complex of phospholipase A2 with a gramine derivative at 2.2 resolution
TO BE PUBLISHED
|
|
2QII
 
 | | Crystal Structure Of tRNA-Guanine Transglycosylase (TGT) From Zymomonas mobilis Complexed With Archaeosine Precursor, Preq0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Brenk, R, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-07-04 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glutamate versus glutamine exchange swaps substrate selectivity in tRNA-guanine transglycosylase: insight into the regulation of substrate selectivity by kinetic and crystallographic studies.
J.Mol.Biol., 374, 2007
|
|
2QJ6
 
 | | Crystal structure analysis of a 14 repeat C-terminal fragment of toxin TcdA in Clostridium difficile | | Descriptor: | Toxin A | | Authors: | Albesa-Jove, D, Bertrand, T, Carpenter, L, Lim, J, Brown, K.A, Fairweather, N. | | Deposit date: | 2007-07-06 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solution and crystal structures of the cell binding domain of toxins TcdA and TcdB from Clostridium difficile
To be Published
|
|
2QM9
 
 | | Troglitazone Bound to Fatty Acid Binding Protein 4 | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, Fatty acid-binding protein, adipocyte, ... | | Authors: | Gillilan, R.E, Ayers, S.D, Noy, N. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Activation of Fatty Acid-binding Protein 4
J.Mol.Biol., 372, 2007
|
|
2QPS
 
 | | "Sugar tongs" mutant Y380A in complex with acarbose | | Descriptor: | Alpha-amylase type A isozyme, CALCIUM ION | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
1R7E
 
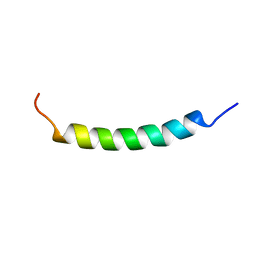 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure. Sample in 100mM SDS). | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R1W
 
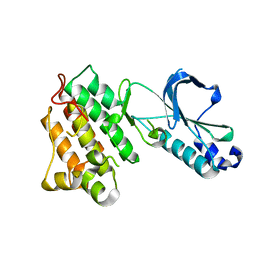 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Schiering, N, Knapp, S, Marconi, M, Flocco, M.M, Cui, J, Perego, R, Rusconi, L, Cristiani, C. | | Deposit date: | 2003-09-25 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1R2C
 
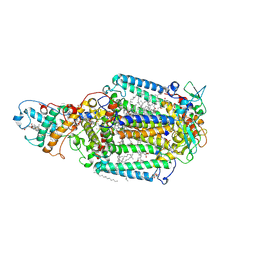 | | PHOTOSYNTHETIC REACTION CENTER BLASTOCHLORIS VIRIDIS (ATCC) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baxter, R.H, Ponomarenko, N, Pahl, R, Srajer, V, Moffat, K, Norris, J.R. | | Deposit date: | 2003-09-26 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Time-resolved crystallographic studies of light-induced structural changes in the photosynthetic reaction center.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1R7S
 
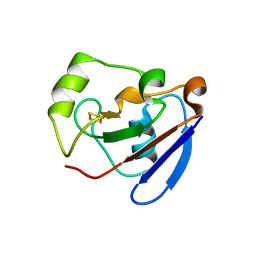 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2QJF
 
 | | Crystal structure of ATP-sulfurylase domain of human PAPS synthetase 1 | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1, POTASSIUM ION | | Authors: | Sekulic, N, Lavie, A. | | Deposit date: | 2007-07-07 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATP-sulfurylase domain of human bifunctional PAPS-synthetase oscillates between dimeric and monomeric forms
To be Published
|
|
4XCU
 
 | | Crystal Structure of FGFR4 with an Irreversible Inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-(2-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl]amino}-3-methylphenyl)propanamide, SULFATE ION | | Authors: | Kim, J.L, Miduturu, C, Hodous, B, Brooijmans, N, Bifulco, N, Guzi, T. | | Deposit date: | 2014-12-18 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | First Selective Small Molecule Inhibitor of FGFR4 for the Treatment of Hepatocellular Carcinomas with an Activated FGFR4 Signaling Pathway.
Cancer Discov, 5, 2015
|
|
6E3S
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist PT2385 | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6EA2
 
 | |
3BJT
 
 | | Pyruvate kinase M2 is a phosphotyrosine binding protein | | Descriptor: | MAGNESIUM ION, OXALATE ION, Pyruvate kinase isozymes M1/M2 | | Authors: | Wu, N. | | Deposit date: | 2007-12-04 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyruvate kinase M2 is a phosphotyrosine-binding protein.
Nature, 452, 2008
|
|
6E3T
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist T1001 | | Descriptor: | (6S)-6-(4-bromophenyl)-2,3,5,6-tetrahydroimidazo[2,1-b][1,3]thiazole, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6EAB
 
 | |
6E3U
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Agonist M1001 | | Descriptor: | 3-{[2-(pyrrolidin-1-yl)phenyl]amino}-1H-1lambda~6~,2-benzothiazole-1,1-dione, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
3AZV
 
 | | Crystal structure of the receptor binding domain | | Descriptor: | D/C mosaic neurotoxin, SULFATE ION | | Authors: | Nuemket, N, Tanaka, Y, Tsukamoto, K, Tsuji, T, Nakamura, K, Kozaki, S, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and mutational analyses of the receptor binding domain of botulinum D/C mosaic neurotoxin: insight into the ganglioside binding mechanism
Biochem.Biophys.Res.Commun., 411, 2011
|
|
6E4B
 
 | |
3AZW
 
 | | Crystal structure of the receptor binding domain | | Descriptor: | D/C mosaic neurotoxin, SULFATE ION | | Authors: | Nuemket, N, Tanaka, Y, Tsukamoto, K, Tsuji, T, Nakamura, K, Kozaki, S, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural and mutational analyses of the receptor binding domain of botulinum D/C mosaic neurotoxin: insight into the ganglioside binding mechanism
Biochem.Biophys.Res.Commun., 411, 2011
|
|
3BHH
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase IIB isoform 1 (CAMK2B) | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II beta chain, [4-({4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-6-(methylamino)pyrimidin-2-yl}amino)phenyl]acetonitrile | | Authors: | Filippakopoulos, P, Rellos, P, Niesen, F, Burgess, N, Bullock, A, Berridge, G, Pike, A.C.W, Ugochukwu, E, Pilka, E.S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Calcium/Calmodulin-Dependent Protein Kinase IIB Isoform 1 (CAMK2B).
To be Published
|
|
6EAA
 
 | |
6EO1
 
 | | The electron crystallography structure of the cAMP-bound potassium channel MloK1 (PCO-refined) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Kowal, J, Biyani, N, Chami, M, Scherer, S, Rzepiela, A, Baumgartner, P, Upadhyay, V, Nimigean, C, Stahlberg, H. | | Deposit date: | 2017-10-08 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (4.5 Å) | | Cite: | High-Resolution Cryoelectron Microscopy Structure of the Cyclic Nucleotide-Modulated Potassium Channel MloK1 in a Lipid Bilayer.
Structure, 26, 2018
|
|
3BX9
 
 | |
3BXJ
 
 | | Crystal Structure of the C2-GAP Fragment of synGAP | | Descriptor: | Ras GTPase-activating protein SynGAP | | Authors: | Pena, V, Hothorn, M, Eberth, A, Kaschau, N, Parret, A, Gremer, L, Bonneau, F, Ahmadian, M.R, Scheffzek, K. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction.
Embo Rep., 9, 2008
|
|
