4DTZ
 
 | | cytochrome P450 BM3h-8C8 MRI sensor bound to dopamine | | Descriptor: | L-DOPAMINE, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 8C8 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
1J1Y
 
 | | Crystal Structure of PaaI from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PaaI protein | | Authors: | Kunishima, N, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
1IWH
 
 | | Crystal Structure of Horse Carbonmonoxyhemoglobin-Bezafibrate Complex at 1.55A Resolution: A Novel Allosteric Binding Site in R-State Hemoglobin | | Descriptor: | 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, CARBON MONOXIDE, Hemoglobin alpha chain, ... | | Authors: | Shibayama, N, Miura, S, Tame, J.R.H, Yonetani, T, Park, S.-Y. | | Deposit date: | 2002-05-15 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Horse Carbonmonoxyhemoglobin-Bezafibrate Complex at 1.55A Resolution. A Novel Allosteric Binding Site in R-State Hemoglobin
J.BIOL.CHEM., 277, 2002
|
|
1IWT
 
 | | Crystal Structure Analysis of Human lysozyme at 113K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IXC
 
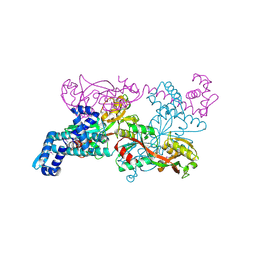 | | Crystal structure of CbnR, a LysR family transcriptional regulator | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-06-18 | | Release date: | 2003-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
2W4Q
 
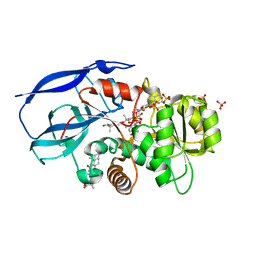 | | Crystal structure of human zinc-binding alcohol dehydrogenase 1 (ZADH1) in ternary complex with NADP and 18beta-glycyrrhetinic acid | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROSTAGLANDIN REDUCTASE 2, ... | | Authors: | Shafqat, N, Yue, W.W, Ugochukwu, E, Picaud, S, Niesen, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2008-11-30 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Zinc-Binding Alcohol Dehydrogenase 1
To be Published
|
|
1J3E
 
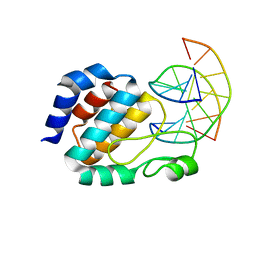 | | Crystal Structure of the E.coli SeqA protein complexed with N6-methyladenine- guanine mismatch DNA | | Descriptor: | 5'-D(*AP*AP*GP*GP*(6MA)P*TP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*AP*GP*CP*CP*TP*T)-3', SeqA protein | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Tanaka, Y, Yamazoe, M, Hiraga, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-24 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein
Nucleic Acids Res., 32, 2004
|
|
2W7L
 
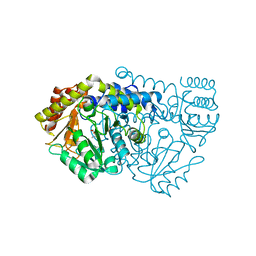 | | Crystal structure of Y61AbsSHMT L-allo-Threonine external aldimine | | Descriptor: | ALLO-THREONINE, PYRIDOXAL-5'-PHOSPHATE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-12-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
1IN3
 
 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
2VSA
 
 | | Structure and mode of action of a mosquitocidal holotoxin | | Descriptor: | MOSQUITOCIDAL TOXIN | | Authors: | Treiber, N, Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure and Mode of Action of a Mosquitocidal Holotoxin
J.Mol.Biol., 381, 2008
|
|
2W7K
 
 | | Crystal structure of Y61AbsSHMT L-Serine external aldimine | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-12-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
2VWL
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID ((3R,5S)-1-{[2-FLUORO-4-(2-OXO-PYRIDIN-1-YL)-PHENYLCARBAMOYL]-METHYL}-5-HYDROXYMETHYL-PYRROLIDIN-3-YL)-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2W2A
 
 | | Crystal Structure of p-coumaric Acid Decarboxylase from Lactobacillus plantarum: structural insights into the active site and decarboxylation catalytic mechanism | | Descriptor: | P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, de las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
1J3Y
 
 | | Direct observation of photolysis-induced tertiary structural changes in human hemoglobin; Crystal structure of alpha(Fe)-beta(Ni) hemoglobin (laser photolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1IRI
 
 | | Crystal structure of human autocrine motility factor complexed with an inhibitor | | Descriptor: | ERYTHOSE-4-PHOSPHATE, autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-10-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
4DCI
 
 | | Crystal structure of unknown funciton protein from Synechococcus sp. WH 8102 | | Descriptor: | SULFATE ION, uncharacterized protein | | Authors: | Chang, C, Marshall, N, Bearden, J, Palenik, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of unknown function protein from Synechococcus sp. WH 8102
To be Published
|
|
1IU5
 
 | | X-ray Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1J9A
 
 | | OLIGORIBONUCLEASE | | Descriptor: | OLIGORIBONUCLEASE, SULFATE ION | | Authors: | Bonander, N, Tordova, M, Ladner, J.E, Eisenstein, E, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-24 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of Haemophilus Influenzae HI1715, an Oligoribonuclease
To be Published
|
|
1IU6
 
 | | Neutron Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.6 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2VPL
 
 | | The structure of the complex between the first domain of L1 protein from Thermus thermophilus and mRNA from Methanococcus jannaschii | | Descriptor: | 50S RIBOSOMAL PROTEIN L1, FRAGMENT OF MRNA FOR L1-OPERON CONTAINING REGULATOR L1-BINDING SITE, POTASSIUM ION | | Authors: | Kljashtorny, V, Tishchenko, S, Nevskaya, N, Nikonov, S, Garber, M. | | Deposit date: | 2008-03-01 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Domain II of Thermus thermophilus ribosomal protein L1 hinders recognition of its mRNA.
J. Mol. Biol., 383, 2008
|
|
1JNH
 
 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | ESTRADIOL-6 CARBOXYL-METHYL-OXIME, monoclonal anti-estradiol 10G6D6 Fab heavy chain, monoclonal anti-estradiol 10G6D6 Fab light chain | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
2VVV
 
 | | Aminopyrrolidine-related triazole Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-1H-1,2,4-triazol-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2W7G
 
 | | Crystal structure of Y51FbsSHMT L-allo-Threonine extrnal aldimine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALLO-THREONINE, PHOSPHATE ION, ... | | Authors: | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-12-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
1JOS
 
 | | Ribosome Binding Factor A(rbfA) | | Descriptor: | RIBOSOME-BINDING FACTOR A | | Authors: | Bonander, N, Tordova, M, Howard, A.J, Eisenstein, E, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-30 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7-A Crystal Structure of HI1288 - Ribosome Binding Factor A (rbfA), a Cold Response Protein
To be Published
|
|
1K25
 
 | |
