1B4A
 
 | | STRUCTURE OF THE ARGININE REPRESSOR FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | ARGININE REPRESSOR | | Authors: | Ni, J, Sakanyan, V, Charlier, D, Glansdorff, N, Van Duyne, G.D. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the arginine repressor from Bacillus stearothermophilus.
Nat.Struct.Biol., 6, 1999
|
|
1AV2
 
 | | Gramicidin A/CsCl complex, active as a dimer | | Descriptor: | CESIUM ION, CHLORIDE ION, GRAMICIDIN A, ... | | Authors: | Burkhart, B.M, Li, N, Langs, D.A, Duax, W.L. | | Deposit date: | 1997-09-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Conducting Form of Gramicidin a is a Right-Handed Double-Stranded Double Helix.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1T29
 
 | | Crystal structure of the BRCA1 BRCT repeats bound to a phosphorylated BACH1 peptide | | Descriptor: | BACH1 phosphorylated peptide, Breast cancer type 1 susceptibility protein | | Authors: | Shiozaki, E.N, Gu, L, Yan, N, Shi, Y. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling.
Mol.Cell, 14, 2004
|
|
1T2X
 
 | | Glactose oxidase C383S mutant identified by directed evolution | | Descriptor: | ACETATE ION, COPPER (II) ION, Galactose Oxidase, ... | | Authors: | Wilkinson, D, Akumanyi, N, Hurtado-Guerrero, R, Dawkes, H, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution.
Protein Eng.Des.Sel., 17, 2004
|
|
2ZIR
 
 | | Crystal Structure of rat protein farnesyltransferase complexed with a benzofuran inhibitor and FPP | | Descriptor: | 2-[(S)-(4-chlorophenyl)(hydroxy)(1-methyl-1H-imidazol-5-yl)methyl]-N-morpholin-4-yl-7-phenyl-1-benzofuran-5-carboxamide, FARNESYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Fukami, T.A, Sogabe, S, Nagata, Y, Kondoh, O, Ishii, N. | | Deposit date: | 2008-02-22 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and structure-activity relationships of novel benzofuran farnesyltransferase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7F0N
 
 | |
1T7S
 
 | | Structural Genomics of Caenorhabditis elegans: Structure of BAG-1 protein | | Descriptor: | BAG-1 cochaperone | | Authors: | Symersky, J, Zhang, Y, Schormann, N, Li, S, Bunzel, R, Pruett, P, Luan, C.-H, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of the BAG domain.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T8B
 
 | | Crystal structure of refolded PHOU-like protein (gi 2983430) from Aquifex aeolicus | | Descriptor: | Phosphate transport system protein phoU homolog | | Authors: | Oganesyan, V, Kim, S.-H, Oganesyan, N, Jancarik, J, Adams, P.D, Kim, R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Crystal structure of the "PhoU-like" phosphate uptake regulator from Aquifex aeolicus.
J.Bacteriol., 187, 2005
|
|
1C4X
 
 | | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE (BPHD) FROM RHODOCOCCUS SP. STRAIN RHA1 | | Descriptor: | PROTEIN (2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE) | | Authors: | Nandhagopal, N, Senda, T, Mitsui, Y. | | Deposit date: | 1999-09-30 | | Release date: | 1999-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Microbial 2-Hydroxyl-6-Oxo-6-Phenylhexa-2,4- Dienoic Acid (Hpda) Hydrolase (Bphd Enzyme) from Rhodococcus Sp. Strain Rha1, in the Pcb Degradation Pathway
Proc.Jpn.Acad.,Ser.B, 73, 1997
|
|
1TD7
 
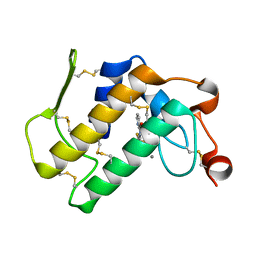 | | Interactions of a specific non-steroidal anti-inflammatory drug (NSAID) with group I phospholipase A2 (PLA2): Crystal structure of the complex formed between PLA2 and niflumic acid at 2.5 A resolution | | Descriptor: | 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Jabeen, T, Singh, N, Singh, R.K, Sharma, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-steroidal anti-inflammatory drugs as potent inhibitors of phospholipase A2: structure of the complex of phospholipase A2 with niflumic acid at 2.5 Angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2ZTN
 
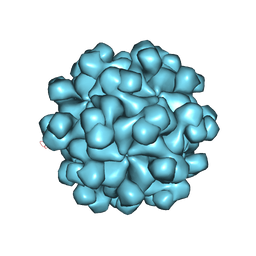 | | Hepatitis E virus ORF2 (Genotype 3) | | Descriptor: | Capsid protein | | Authors: | Yamashita, T, Unno, H, Mori, Y, Li, T.C, Takeda, N, Matsuura, Y. | | Deposit date: | 2008-10-08 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1Q5T
 
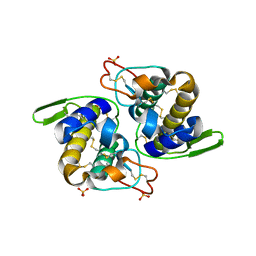 | | Gln48 PLA2 separated from Vipoxin from the venom of Vipera ammodytes meridionalis. | | Descriptor: | Phospholipase A2 inhibitor, SULFATE ION | | Authors: | Georgieva, D.N, Perbandt, M, Rypniewski, W, Hristov, K, Genov, N, Betzel, C. | | Deposit date: | 2003-08-11 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray structure of a snake venom Gln48 phospholipase A2 at 1.9A resolution reveals
anion-binding sites.
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1PX9
 
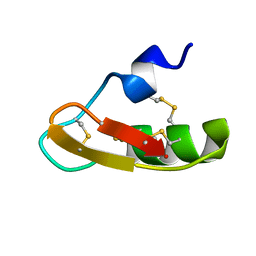 | | Solution structure of the native CnErg1 Ergtoxin, a highly specific inhibitor of HERG channel | | Descriptor: | ergtoxin | | Authors: | Frenal, K, Wecker, K, Gurrola, G.B, Possani, L.D, Wolff, N, Delepierre, M. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Exploring structural features of the interaction between the scorpion toxinCnErg1 and ERG K+ channels.
Proteins, 56, 2004
|
|
1C8Q
 
 | |
2ZLA
 
 | | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation and crystal structure | | Descriptor: | (1R,2S,3R,5Z,7E)-17-{(1R)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethoxy)-9,10-secoestra-5,7,16-triene-1,3-diol, Coactivator peptide DRIP, Vitamin D3 receptor | | Authors: | Shimizu, M, Miyamoto, Y, Nakabayashi, M, Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure
Bioorg.Med.Chem., 16, 2008
|
|
1PZV
 
 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1Q01
 
 | | Lebetin peptides, a new class of potent aggregation inhibitors | | Descriptor: | lebetin 2 isoform alpha | | Authors: | Mosbah, A, Marrakchi, N, Ganzalez, M.J, Van Rietschoten, J, Giralt, E, El Ayeb, M, Rochat, H, Sabatier, J.M, Darbon, H, Mabrouk, K. | | Deposit date: | 2003-07-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Lebetin peptides, a new class of potent aggregation inhibitors
To be Published
|
|
1BZ8
 
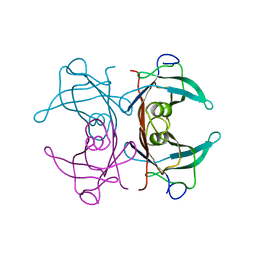 | |
1Q3N
 
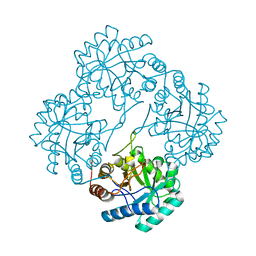 | | Crystal structure of KDO8P synthase in its binary complex with substrate PEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHOENOLPYRUVATE | | Authors: | Vainer, R, Belakhov, V, Rabkin, E, Baasov, T, Adir, N. | | Deposit date: | 2003-07-31 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Escherichia coli KDO8P synthase complexes reveal the source of catalytic irreversibility.
J.Mol.Biol., 351, 2005
|
|
7FCM
 
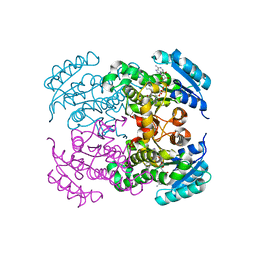 | | Crystal structure of Moraxella catarrhalis enoyl-ACP-reductase (FabI) in complex with NAD and Triclosan | | Descriptor: | CALCIUM ION, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Katiki, M, Neetu, N, Pratap, S, Kumar, P. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Biochemical and structural basis for Moraxella catarrhalis enoyl-acyl carrier protein reductase (FabI) inhibition by triclosan and estradiol.
Biochimie, 198, 2022
|
|
1QOJ
 
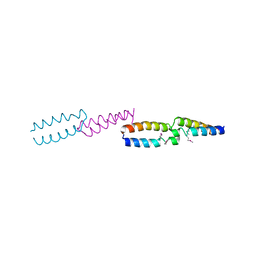 | | Crystal Structure of E.coli UvrB C-terminal domain, and a model for UvrB-UvrC interaction. | | Descriptor: | UVRB | | Authors: | Sohi, M, Alexandrovich, A, Moolenaar, G, Visse, R, Goosen, N, Vernede, X, Fontecilla-Camps, J, Champness, J, Sanderson, M.R. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-10 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of E.Coli Uvrb C-Terminal Domain, and a Model for Uvrb-Uvrc Interaction
FEBS Lett., 465, 2000
|
|
1BQJ
 
 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Yang, C, Wang, S, Sutherland, C, Sinha, N, Kang, C.H. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1QOX
 
 | |
3A20
 
 | |
1QVP
 
 | | C terminal SH3-like domain from Diphtheria toxin Repressor residues 144-226. | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Marin, V, Bhattacharya, N, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
