6WEX
 
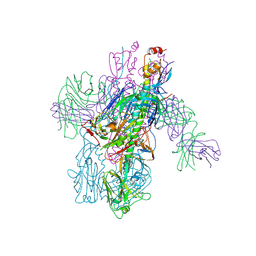 | | Crystal Structure of Broadly Neutralizing Antibody 3I14-D93N Mutant Bound to the Influenza A H6 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
6WOV
 
 | | Cryo-EM structure of recombinant mouse Ryanodine Receptor type 2 wild type in complex with FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Nayak, A.R, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
7QD8
 
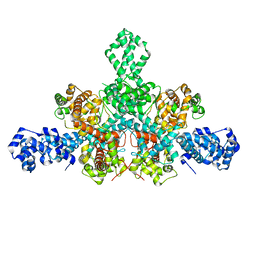 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in apo state | | Descriptor: | Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD4
 
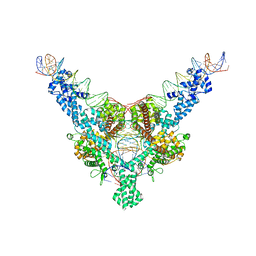 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 100 bp long transposon end DNA | | Descriptor: | IR100 DNA substrate, none transferred strand, transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
8FMC
 
 | |
8FMD
 
 | |
8FOQ
 
 | |
8FME
 
 | |
6WV5
 
 | | Human VKOR C43S mutant with vitamin K1 epoxide | | Descriptor: | (2R,3R)-2-hydroxy-3-methyl-2-[(2E,7S)-3,7,11,15-tetramethylhexadec-2-en-1-yl]-2,3-dihydronaphthalene-1,4-dione, Vitamin K epoxide reductase Cys43Ser mutant, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
8FOS
 
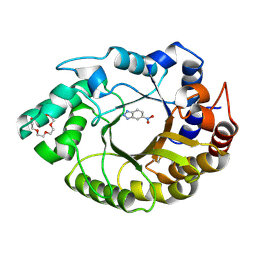 | |
8FW2
 
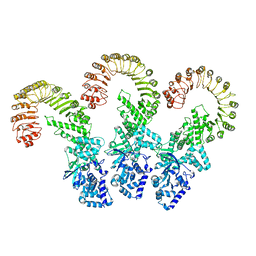 | | Cryo-EM structure of full-length human NLRC4 inflammasome with C11 symmetry | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Matico, R.E, Yu, X, Miller, R, Somani, S, Ricketts, M.D, Kumar, N, Steele, R.A, Medley, Q, Berger, S, Faustin, B, Sharma, S. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of the human NAIP/NLRC4 inflammasome assembly and pathogen sensing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FVU
 
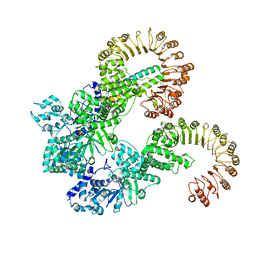 | | Cryo-EM structure of human Needle/NAIP/NLRC4 (R288A) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1, Lethal factor,Type III secretion system protein, ... | | Authors: | Matico, R.E, Yu, X, Miller, R, Somani, S, Ricketts, M.D, Kumar, N, Steele, R.A, Medley, Q, Berger, S, Faustin, B, Sharma, S. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the human NAIP/NLRC4 inflammasome assembly and pathogen sensing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FW9
 
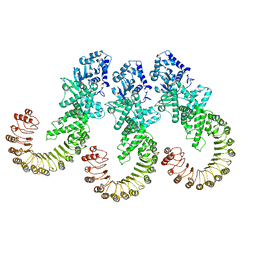 | | Cryo-EM structure of full-length human NLRC4 inflammasome with C12 symmetry | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Matico, R.E, Yu, X, Miller, R, Somani, S, Ricketts, M.D, Kumar, N, Steele, R.A, Medley, Q, Berger, S, Faustin, B, Sharma, S. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Structural basis of the human NAIP/NLRC4 inflammasome assembly and pathogen sensing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6WHR
 
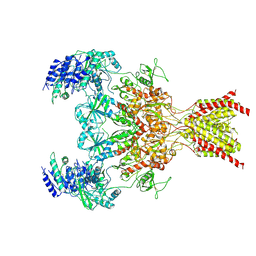 | | GluN1b-GluN2B NMDA receptor in non-active 2 conformation at 4 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
7QD6
 
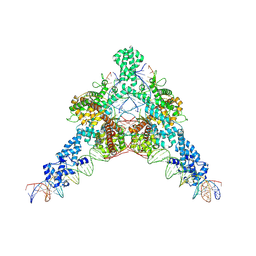 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with strand-transfer like DNA product | | Descriptor: | IR71st non transferred strand, IR71st transferred strand, Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD5
 
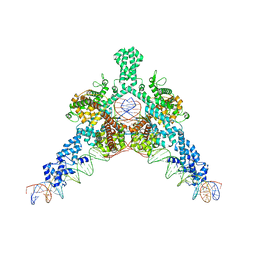 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 48 bp long transposon end DNA | | Descriptor: | IR48 DNA substrate, non transferred strand, IR48 transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
6WCB
 
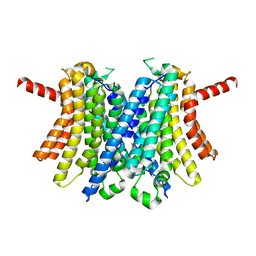 | | Human open state TMEM175 in CsCl | | Descriptor: | CESIUM ION, Endosomal/lysosomal potassium channel TMEM175 | | Authors: | Oh, S, Paknejad, N, Hite, R.K. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Gating and selectivity mechanisms for the lysosomal K + channel TMEM175.
Elife, 9, 2020
|
|
7QDA
 
 | | Crystal structure of CalpL | | Descriptor: | CalpL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
6WCY
 
 | | N160D Deamidation Mutant of Human gammaD-Crystallin | | Descriptor: | Gamma-crystallin D, SULFATE ION | | Authors: | Whitley, M.J, Rathi, N, Ambarian, M, Gronenborn, A.M. | | Deposit date: | 2020-03-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.204 Å) | | Cite: | Assessing the Structures and Interactions of gamma D-Crystallin Deamidation Variants.
Structure, 29, 2021
|
|
1R4S
 
 | | URATE OXIDASE FROM ASPERGILLUS FLAVUS COMPLEXED WITH ITS INHIBITOR 9-METHYL URIC ACID | | Descriptor: | 9-METHYL URIC ACID, CYSTEINE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Prange, T. | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6WHX
 
 | | GluN1b-GluN2B NMDA receptor in complex with GluN2B antagonist SDZ 220-040, class 2 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ionotropic glutamate receptor , ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
7Q94
 
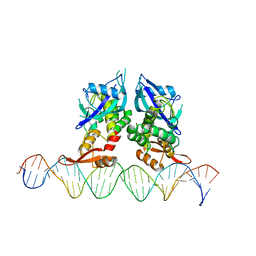 | | Crystal Structure of Agrobacterium tumefaciens NADQ, DNA complex. | | Descriptor: | DNA binding region (31-MER), NADQ transcription factor | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
6WKP
 
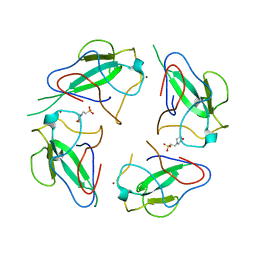 | | Crystal structure of RNA-binding domain of nucleocapsid phosphoprotein from SARS CoV-2, monoclinic crystal form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7Q91
 
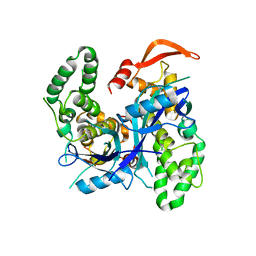 | | Crystal Structure of Agrobacterium tumefaciens NADQ, native form. | | Descriptor: | NADQ transcription factor, SODIUM ION | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
6WH7
 
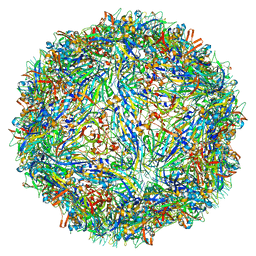 | | Capsid structure of Penaeus monodon metallodensovirus following EDTA treatment | | Descriptor: | Penaeus monodon metallodensovirus major capsid protein | | Authors: | Penzes, J.J, Pham, H.T, Chipman, P, Bhattacharya, N, McKenna, R, Agbandje-McKenna, M, Tijssen, P. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Molecular biology and structure of a novel penaeid shrimp densovirus elucidate convergent parvoviral host capsid evolution.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
