5IX6
 
 | | X-ray structure of Auoxo3-encapsulated horse spleen apoferritin | | Descriptor: | CADMIUM ION, Ferritin light chain, GOLD ION, ... | | Authors: | Ferraro, G, Pontillo, N, Merlino, A. | | Deposit date: | 2016-03-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Gold-based drug encapsulation within a ferritin nanocage: X-ray structure and biological evaluation as a potential anticancer agent of the Auoxo3-loaded protein.
Chem.Commun.(Camb.), 52, 2016
|
|
5IXB
 
 | | Structure of human Melanoma Inhibitory Activity (MIA) Protein in complex with Pyrimidin-2-amine | | Descriptor: | ACETATE ION, Melanoma-derived growth regulatory protein, PYRIMIDIN-2-AMINE | | Authors: | Yip, K.T, Gasper, R, Zhong, X.Y, Seibel, N, Puetz, S, Autzen, J, Scherkenbeck, J, Hofmann, E, Stoll, R. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Small Molecules Antagonise the MIA-Fibronectin Interaction in Malignant Melanoma.
Sci Rep, 6, 2016
|
|
8BF2
 
 | |
5J83
 
 | |
5LKR
 
 | | Human Butyrylcholinesterase complexed with N-Propargyliperidines | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Coquelle, N, Knez, D, Colletier, J.P, Gobec, S. | | Deposit date: | 2016-07-23 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | N-Propargylpiperidines with naphthalene-2-carboxamide or naphthalene-2-sulfonamide moieties: Potential multifunctional anti-Alzheimer's agents.
Bioorg. Med. Chem., 25, 2017
|
|
5LOB
 
 | |
5LOW
 
 | |
5M4V
 
 | | X-ray structure of the mambaquaretin-1, a selective antagonist of the vasopressin type 2 receptor | | Descriptor: | CHLORIDE ION, Mambaquaretin-1, S-1,2-PROPANEDIOL | | Authors: | Stura, E.A, Vera, L, Ciolek, J, Mourier, G, Gilles, N. | | Deposit date: | 2016-10-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LCM
 
 | | STRUCTURE OF the RAD14 DNA-binding domain IN COMPLEX WITH N2-acetylaminonaphtyl- GUANINE CONTAINING DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*TP*AP*CP*(AAN)P*TP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*G)-3'), DNA repair protein RAD14, ... | | Authors: | Schneider, S, Ebert, C, Simon, N, Carell, T. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Chembiochem, 18, 2017
|
|
5LHE
 
 | |
5LLU
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with L-ASP | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, SODIUM ION | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-28 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
5LZA
 
 | | Structure of the 70S ribosome with SECIS-mRNA and P-site tRNA (Initial complex, IC) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LM7
 
 | | Crystal structure of the lambda N-Nus factor complex | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | Authors: | Said, N, Santos, K, Weber, G, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
7Z58
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound Calpeptin | | Descriptor: | 1,2-ETHANEDIOL, Calpeptin, Cathepsin L, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
5LMZ
 
 | | Fluorinase from Streptomyces sp. MA37 | | Descriptor: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | Authors: | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
5LZF
 
 | | Structure of the 70S ribosome with fMetSec-tRNASec in the hybrid pre-translocation state (H) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LGO
 
 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 15 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)-3-methoxy-phenyl]carbonyl-4-[4-(2-cyclopropylethoxy)-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidin-7-yl]-~{N}-methyl-pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5ELJ
 
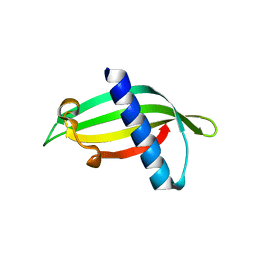 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SUMO-Affirmer-S2D5, Small ubiquitin-related modifier 1 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISI
 
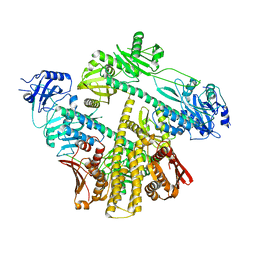 | | Photochromobilin-free form of Arabidopsis thaliana phytochrome A - apo-AtphyA | | Descriptor: | Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
4OAQ
 
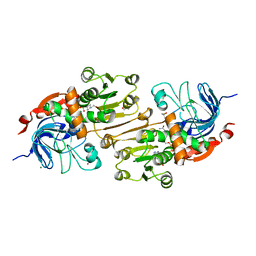 | | Crystal structure of the R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Aggrawal, N, Mandal, P.K, Gautham, N, Chadha, A. | | Deposit date: | 2014-01-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Insights into the stereoselectivity of R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330: Biochemical Characterization and Crystal structure studies
To be Published
|
|
2P6L
 
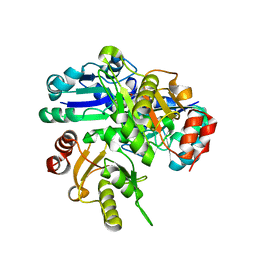 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Matsuura, Y, Ono, N, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2P8U
 
 | | Crystal structure of human 3-hydroxy-3-methylglutaryl CoA synthase I | | Descriptor: | COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Turnbull, A, Shafqat, N, Salah, E, Niesen, F.H, Burgess, N, Bunkoczi, G, Debreczeni, J, Pike, A.C.W, Umeano, C, Gorrec, F, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human HMG-CoA synthase isoforms provide insights into inherited ketogenesis disorders and inhibitor design.
J.Mol.Biol., 398, 2010
|
|
2P6K
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, diphthine synthase | | Authors: | Yamamoto, H, Matsuura, Y, Ono, N, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-18 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
