7VHP
 
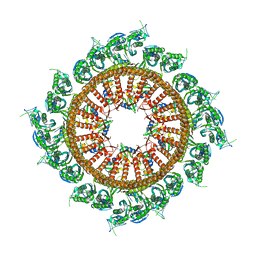 | | Structural insights into the membrane microdomain organization by SPFH family proteins | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Protein HflK | | Authors: | Ma, C.Y, Wang, C.K, Luo, D.Y, Yan, L, Yang, W.X, Li, N.N, Gao, N. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the membrane microdomain organization by SPFH family proteins.
Cell Res., 32, 2022
|
|
1OHJ
 
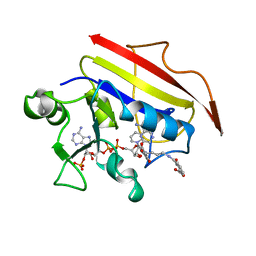 | | HUMAN DIHYDROFOLATE REDUCTASE, MONOCLINIC (P21) CRYSTAL FORM | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1997-09-17 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes reduced with nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523.
Biochemistry, 36, 1997
|
|
1OHK
 
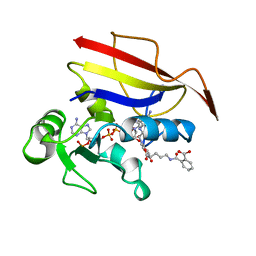 | | HUMAN DIHYDROFOLATE REDUCTASE, ORTHORHOMBIC (P21 21 21) CRYSTAL FORM | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1997-09-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes reduced with nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523.
Biochemistry, 36, 1997
|
|
4U63
 
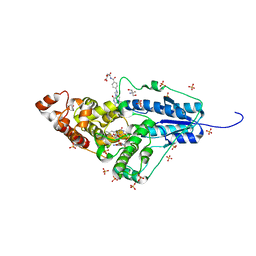 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
5ZR1
 
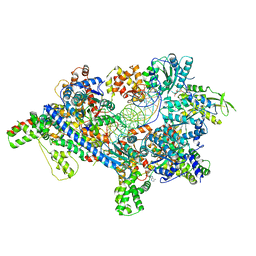 | | Saccharomyces Cerevisiae Origin Recognition Complex Bound to a 72-bp Origin DNA containing ACS and B1 element | | Descriptor: | 72bp-oring DNA, ACS305, A-rich, ... | | Authors: | Li, N, Lam, W.H, Zhai, Y, Cheng, J, Cheng, E, Zhao, Y, Gao, N, Tye, B.K. | | Deposit date: | 2018-04-21 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the origin recognition complex bound to DNA replication origin.
Nature, 559, 2018
|
|
5ZRV
 
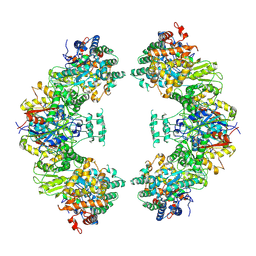 | | Structure of human mitochondrial trifunctional protein, octamer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1MIE
 
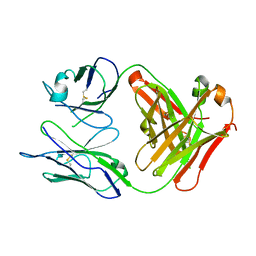 | | Crystal Structure Of The Fab Fragment of Esterolytic Antibody MS5-393 | | Descriptor: | IMMUNOGLOBULIN MS5-393 | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-23 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1LY4
 
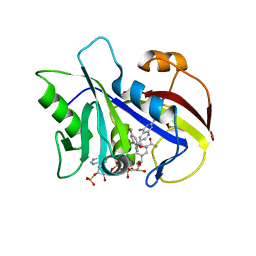 | | Analysis of quinazoline and PYRIDO[2,3D]PYRIMIDINE N9-C10 reversed bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5ZJA
 
 | | human D-amino acid oxidase complexed with 5-chlorothiophene-2-carboxylic acid | | Descriptor: | 5-chloro thiophene-2-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
6OPV
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPZ
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, I54L, A71V, L76V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
1PMY
 
 | | REFINED CRYSTAL STRUCTURE OF PSEUDOAZURIN FROM METHYLOBACTERIUM EXTORQUENS AM1 AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Kai, Y, Harada, S, Kasai, N, Ohshiro, Y, Suzuki, S, Kohzuma, T, Tobari, J. | | Deposit date: | 1994-01-28 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Refined crystal structure of pseudoazurin from Methylobacterium extorquens AM1 at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PR9
 
 | | Human L-Xylulose Reductase Holoenzyme | | Descriptor: | DIHYDROGENPHOSPHATE ION, L-XYLULOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | El-Kabbani, O, Ishikura, S, Darmanin, C, Carbone, V, Chung, R.P.-T, Usami, N, Hara, A. | | Deposit date: | 2003-06-20 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of human L-xylulose reductase holoenzyme: probing the role of Asn107 with site-directed mutagenesis
Proteins, 55, 2004
|
|
1Q3Y
 
 | | NMR structure of the Cys28His mutant (D form) of the nucleocapsid protein NCp7 of HIV-1. | | Descriptor: | GAG protein, ZINC ION | | Authors: | Ramboarina, S, Druillennec, S, Morellet, N, Bouaziz, S, Roques, B.P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Target specificity of human immunodeficiency virus type 1 NCp7 requires an intact conformation of its CCHC N-terminal zinc finger.
J.Virol., 78, 2004
|
|
1Q3Z
 
 | | NMR structure of the Cys28His mutant (E form) of the nucleocapsid protein NCp7 of HIV-1. | | Descriptor: | GAG protein, ZINC ION | | Authors: | Ramboarina, S, Druillennec, S, Morellet, N, Bouaziz, S, Roques, B.P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Target specificity of human immunodeficiency virus type 1 NCp7 requires an intact conformation of its CCHC N-terminal zinc finger.
J.Virol., 78, 2004
|
|
1MJ7
 
 | | Crystal Structure Of The Complex Of The Fab fragment of Esterolytic Antibody MS5-393 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS5-393, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MO5
 
 | |
1MO4
 
 | |
6N7V
 
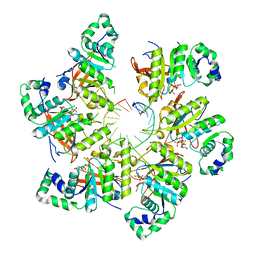 | | Structure of bacteriophage T7 gp4 (helicase-primase, E343Q mutant) in complex with ssDNA, dTTP, AC dinucleotide, and CTP (from multiple lead complexes) | | Descriptor: | DNA (93-MER), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6OOT
 
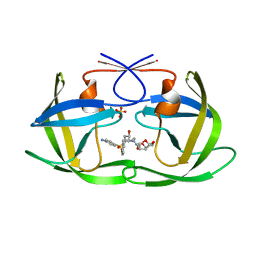 | | HIV-1 Protease NL4-3 L89V, L90M Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
7UTF
 
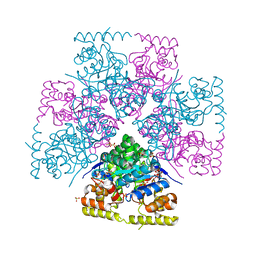 | | Structure-Function characterization of an aldo-keto reductase involved in detoxification of the mycotoxin, deoxynivalenol | | Descriptor: | CITRATE ANION, Putative oxidoreductase, aryl-alcohol dehydrogenase like protein, ... | | Authors: | Abraham, N, Schroeter, K.L, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function characterization of an aldo-keto reductase involved in detoxification of the mycotoxin, deoxynivalenol.
Sci Rep, 12, 2022
|
|
8IVQ
 
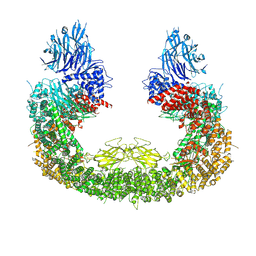 | | Cryo-EM structure of mouse BIRC6, Global map | | Descriptor: | Isoform 2 of Baculoviral IAP repeat-containing protein 6 | | Authors: | Liu, S, Jiang, T, Bu, F, Zhao, J, Wang, G, Li, N, Gao, N, Qiu, X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy.
Nat Commun, 15, 2024
|
|
6OH1
 
 | | IgA1 Protease G5 domain structure | | Descriptor: | Immunoglobulin A1 protease | | Authors: | Eisenmesser, E.Z, Chi, Y.C, Paukovich, N, Redzic, J.S, Rahkola, J.T, Janoff, E.N. | | Deposit date: | 2019-04-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Streptococcus pneumoniae G5 domains bind different ligands.
Protein Sci., 28, 2019
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
1MJJ
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 AND A TRANSITION-STATE ANALOG | | Descriptor: | IMMUNOGLOBULIN MS6-12, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
