5H5P
 
 | |
4C3Y
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-KETOSTEROID DEHYDROGENASE, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rohman, A, van Oosterwijk, N, Thunnissen, A.M.W.H, Dijkstra, B.W. | | Deposit date: | 2013-08-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis of 3-Ketosteroid Delta1-Dehydrogenase from Rhodococcus Erythropolis Sq1 Explain its Catalytic Mechanism
J.Biol.Chem., 288, 2013
|
|
5H6R
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kkuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
6AW4
 
 | | 1.50A resolution structure of catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catechol O-methyltransferase, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
2V2R
 
 | | Mutant (E53,56,57,60Q and R59M) recombinant horse spleen apoferritin cocrystallized with haemin in basic conditions | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN, GLYCEROL, ... | | Authors: | De Val, N, Declercq, J.P. | | Deposit date: | 2007-06-06 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Haemin Demetallation by L-Chain Apoferritins
J.Inorg.Biochem., 112, 2012
|
|
5HFN
 
 | |
2V3K
 
 | | The yeast ribosome synthesis factor Emg1 alpha beta knot fold methyltransferase | | Descriptor: | ESSENTIAL FOR MITOTIC GROWTH 1, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Leulliot, N, Bohnsack, M.T, Graille, M, Tollervey, D, VanTilbeurgh, H. | | Deposit date: | 2007-06-18 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Yeast Ribosome Synthesis Factor Emg1 is a Novel Member of the Superfamily of Alpha/Beta Knot Fold Methyltransferases.
Nucleic Acids Res., 36, 2008
|
|
4C3X
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 | | Descriptor: | 3-KETOSTEROID DEHYDROGENASE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rohman, A, van Oosterwijk, N, Thunnissen, A.M.W.H, Dijkstra, B.W. | | Deposit date: | 2013-08-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis of 3-Ketosteroid Delta1-Dehydrogenase from Rhodococcus Erythropolis Sq1 Explain its Catalytic Mechanism
J.Biol.Chem., 288, 2013
|
|
5GJ9
 
 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with POA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRAZINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
4CD3
 
 | | RnNTPDase2 X4 variant in complex with PSB-071 | | Descriptor: | 1-AMINO-4-(3-METHYLPHENYL)AMINO-9,10-DIOXO-9,10-DIHYDROANTHRACENE-2-SULFONATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, GLYCEROL | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2013-10-29 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Ntpdase2 in Complex with the Sulfoanthraquinone Inhibitor Psb-071.
J.Struct.Biol., 185, 2014
|
|
5ZJR
 
 | | Structure of AbdB/Exd complex bound to a 'Magenta14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*TP*CP*GP*TP*AP*AP*AP*TP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*AP*TP*TP*TP*AP*CP*GP*AP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Zeiske, T, Baburajendran, N, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
5G32
 
 | | Structure of Rad14 in complex with acetylaminophenyl-guanine containing DNA | | Descriptor: | 5'-D(*GP*CP*TP*CP*TP*AP*6FKP*TP*CP*AP*TP*CP*AP*CP)-3', 5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*GP)-3', RAD14, ... | | Authors: | Simon, N, Ebert, C, Schneider, S. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Bulky Adduct DNA Lesion Recognition by the Nucleotide Excision Repair Protein Rad14.
Chemistry, 22, 2016
|
|
5GO9
 
 | |
5G59
 
 | |
2VMU
 
 | | Crystal structure of Y60AbsSHMT crystallized in the presence of L- allo-Thr | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, PHOSPHATE ION, ... | | Authors: | Rajaram, V, Pai, V.R, Bisht, S, Bhavani, B.S, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-01-29 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and Functional Studies of Bacillus Stearothermophilus Serine Hydroxymethyltransferase: The Role of Asn(341), Tyr(60) and Phe(351) in Tetrahydrofolate Binding.
Biochem.J., 418, 2009
|
|
5GGG
 
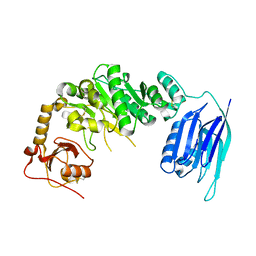 | |
5GJW
 
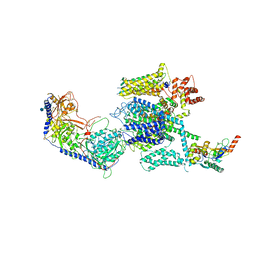 | | Structure of the mammalian voltage-gated calcium channel Cav1.1 complex for ClassII map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Yan, Z, Li, Z.Q, Zhou, Q, Yan, N. | | Deposit date: | 2016-07-02 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Nature, 537, 2016
|
|
5GGN
 
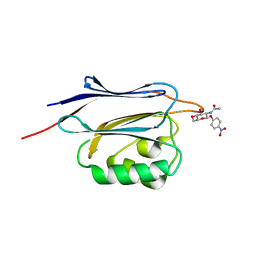 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5Z30
 
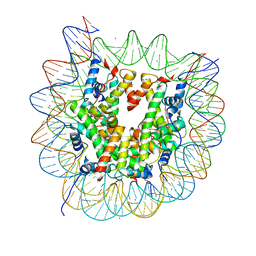 | | The crystal structure of the nucleosome containing a cancer-associated histone H2A.Z R80C mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.Z, ... | | Authors: | Horikoshi, N, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2018-01-05 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
2PMT
 
 | | GLUTATHIONE TRANSFERASE FROM PROTEUS MIRABILIS | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Polekhina, G, Feil, S.C, Allocati, N, Masulli, M, Diilio, C, Parker, M.W. | | Deposit date: | 1998-04-28 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.
Structure, 6, 1998
|
|
5GLH
 
 | | Human endothelin receptor type-B in complex with ET-1 | | Descriptor: | Endothelin Receptor Subtype-B, Peptide from Endothelin-1 | | Authors: | Shihoya, W, Nishizawa, T, Okuta, A, Tani, K, Fujiyoshi, Y, Dohmae, N, Nureki, O, Doi, T. | | Deposit date: | 2016-07-11 | | Release date: | 2016-09-07 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation mechanism of endothelin ETB receptor by endothelin-1.
Nature, 537, 2016
|
|
5GLQ
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with l-arabinose and xylotriose, calcium-free form | | Descriptor: | Glycoside hydrolase family 43, SODIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5Z6M
 
 | |
5ZB9
 
 | | Crystal structure of Rtt109 from Aspergillus fumigatus | | Descriptor: | DNA damage response protein Rtt109, putative, GLYCEROL | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
2QF8
 
 | | Crystal structure of the complex of Buffalo Secretory Glycoprotein with tetrasaccharide at 2.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Singh, A.K, Jain, R, Sinha, M, Kumar, A, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the complex of Buffalo Secretory Glycoprotein with Tetrasaccharide at 2.8A resolution
To be Published
|
|
