6SZS
 
 | | Release factor-dependent ribosome rescue by BrfA in the Gram-positive bacterium Bacillus subtilis | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Muller, C, Beckert, B, Wilson, D.N. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Release factor-dependent ribosome rescue by BrfA in the Gram-positive bacterium Bacillus subtilis.
Nat Commun, 10, 2019
|
|
1XBR
 
 | | T DOMAIN FROM XENOPUS LAEVIS BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP*AP*GP*GP*TP*G P*TP*GP*AP*AP*AP* TP*T)-3'), PROTEIN (T PROTEIN) | | Authors: | Muller, C.W. | | Deposit date: | 1997-07-16 | | Release date: | 1998-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor.
Nature, 389, 1997
|
|
8Q71
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GC-67 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-(4-methoxypyridin-3-yl)carbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Weisse, R.H, Useini, A, Gao, S, Song, L, Liu, Z, Zhan, P. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Trisubstituted Piperazine Derivatives as Noncovalent Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors with Improved Antiviral Activity and Favorable Druggability.
J.Med.Chem., 66, 2023
|
|
8RLN
 
 | | Crystal structure of human adenosine A2A receptor (construct A2A-PSB2-bRIL) complexed with the partial antagonist LUF5834 at the orthosteric pocket | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-azanyl-4-(4-hydroxyphenyl)-6-(1~{H}-imidazol-2-ylmethylsulfanyl)pyridine-3,5-dicarbonitrile, ... | | Authors: | Strater, N, Claff, T, Weisse, R.H, Muller, C.E. | | Deposit date: | 2024-01-03 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Insights into Partial Activation of the Prototypic G Protein-Coupled Adenosine A 2A Receptor.
Acs Pharmacol Transl Sci, 7, 2024
|
|
9IK1
 
 | | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kim, S, Kim, G.R, Kim, Y.O, Han, X, Nagel, J, Kim, J, Song, D.I, Muller, C.E, Yoon, M.H, Jin, M.S, Kim, Y.C. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy.
J.Med.Chem., 67, 2024
|
|
7PYR
 
 | | Crystal structure of the adenosine A2A receptor (A2A-PSB1-bRIL) in complex with preladenant conjugate PSB-2115 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[2-[2-[2-[2-[2-[4-[4-[2-[7-azanyl-4-(furan-2-yl)-3,5,6,8,10,11-hexazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2,4,7,11-pentaen-10-yl]ethyl]piperazin-1-yl]phenoxy]ethanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]-~{N}-[5-[2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9,11-tetraen-8-yl]pentyl]ethanamide, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Claff, T, Klapschinski, T.A, Tiruttani Subhramanyam, U.K, Vaassen, V.J, Schlegel, J.G, Vielmuth, C, Voss, J.H, Labahn, J, Muller, C.E. | | Deposit date: | 2021-10-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Single Stabilizing Point Mutation Enables High-Resolution Co-Crystal Structures of the Adenosine A 2A Receptor with Preladenant Conjugates.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7PX4
 
 | | Crystal structure of the adenosine A2A receptor (A2A-PSB1-bRIL) in complex with preladenant conjugate PSB-2113 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Claff, T, Klapschinski, T.A, Tiruttani Subhramanyam, U.K, Vaassen, V.J, Schlegel, J.G, Vielmuth, C, Voss, J.H, Labahn, J, Muller, C.E. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Single Stabilizing Point Mutation Enables High-Resolution Co-Crystal Structures of the Adenosine A 2A Receptor with Preladenant Conjugates.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6PT2
 
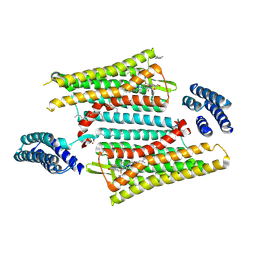 | | Crystal structure of the active delta opioid receptor in complex with the peptide agonist KGCHM07 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Delta opioid receptor, ... | | Authors: | Claff, T, Yu, J, Blais, V, Patel, N, Martin, C, Wu, L, Han, G.W, Holleran, B.J, Van der Poorten, O, Hanson, M.A, Sarret, P, Gendron, L, Cherezov, V, Katritch, V, Ballet, S, Liu, Z, Muller, C.E, Stevens, R.C. | | Deposit date: | 2019-07-14 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elucidating the active delta-opioid receptor crystal structure with peptide and small-molecule agonists.
Sci Adv, 5, 2019
|
|
6PT3
 
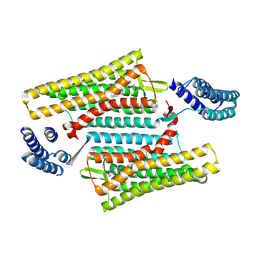 | | Crystal structure of the active delta opioid receptor in complex with the small molecule agonist DPI-287 | | Descriptor: | 4-[(R)-[(2S,5R)-4-benzyl-2,5-dimethylpiperazin-1-yl](3-hydroxyphenyl)methyl]-N,N-diethylbenzamide, Delta opioid receptor | | Authors: | Claff, T, Yu, J, Blais, V, Patel, N, Martin, C, Wu, L, Han, G.W, Holleran, B.J, Van der Poorten, O, Hanson, M.A, Sarret, P, Gendron, L, Cherezov, V, Katritch, V, Ballet, S, Liu, Z, Muller, C.E, Stevens, R.C. | | Deposit date: | 2019-07-14 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Elucidating the active delta-opioid receptor crystal structure with peptide and small-molecule agonists.
Sci Adv, 5, 2019
|
|
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
5AEM
 
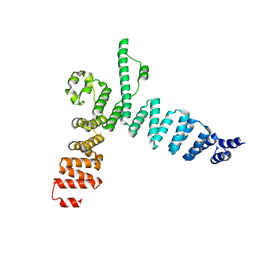 | | Structure of t131 N-terminal TPR array | | Descriptor: | TRANSCRIPTION FACTOR TAU 131 KDA SUBUNIT | | Authors: | Taylor, N.M.I, Muller, C.W. | | Deposit date: | 2015-01-05 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly.
Nat Commun, 6, 2015
|
|
3NSU
 
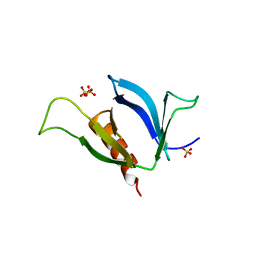 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
6YJ6
 
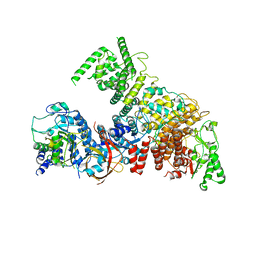 | | Structure of the TFIIIC subcomplex tauA | | Descriptor: | Transcription factor tau 131 kDa subunit, Transcription factor tau 55 kDa subunit, Transcription factor tau 95 kDa subunit,Transcription factor tau 95 kDa subunit | | Authors: | Vorlaender, M.K, Muller, C.W. | | Deposit date: | 2020-04-02 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the TFIIIC subcomplex tau A provides insights into RNA polymerase III pre-initiation complex formation.
Nat Commun, 11, 2020
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
1BG1
 
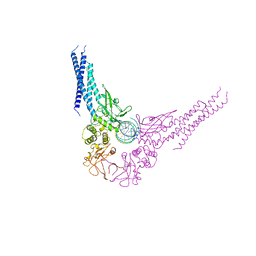 | | TRANSCRIPTION FACTOR STAT3B/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR STAT3B) | | Authors: | Becker, S, Groner, B, Muller, C.W. | | Deposit date: | 1998-06-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Three-dimensional structure of the Stat3beta homodimer bound to DNA.
Nature, 394, 1998
|
|
6HLS
 
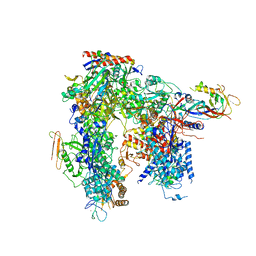 | | Yeast apo RNA polymerase I* | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HLQ
 
 | | Yeast RNA polymerase I* elongation complex bound to nucleotide analog GMPCPP | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HKO
 
 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-07 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HLR
 
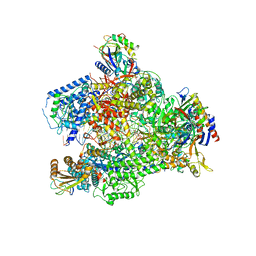 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP (core focused) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
5FJ9
 
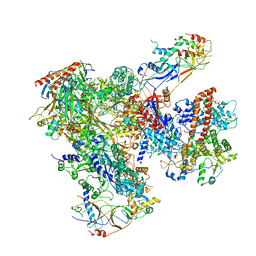 | | Cryo-EM structure of yeast apo RNA polymerase III at 4.6 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
1A3Q
 
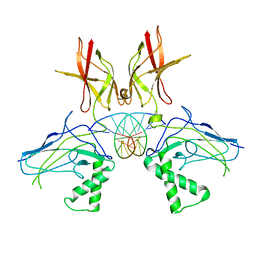 | | HUMAN NF-KAPPA-B P52 BOUND TO DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*CP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B P52) | | Authors: | Cramer, P, Larson, C.J, Verdine, G.L, Muller, C.W. | | Deposit date: | 1998-01-23 | | Release date: | 1998-06-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution.
EMBO J., 16, 1997
|
|
5FJ8
 
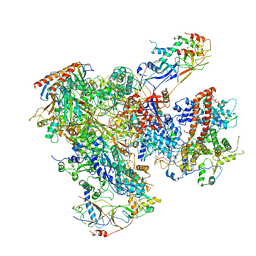 | | Cryo-EM structure of yeast RNA polymerase III elongation complex at 3. 9 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
6T33
 
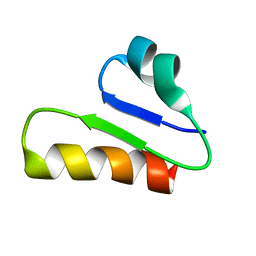 | | The unusual structure of Ruminococcin C1 antimicrobial peptide confers activity against clinical pathogens | | Descriptor: | Ruminococcin C | | Authors: | Chiumento, S, Roblin, C, Bornet, O, Nouailler, M, Muller, C, Basset, C, Kieffer-Jaquinod, S, Coute, Y, Torelli, S, Le Pape, L, Shunemann, V, Jeannot, K, Nicoletti, C, Iranzo, O, Maresca, M, Giardina, T, Fons, M, Devillard, E, Perrier, J, Atta, M, Guerlesquin, F, Lafond, M, Duarte, V. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The unusual structure of Ruminococcin C1 antimicrobial peptide confers clinical properties.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5FJA
 
 | | Cryo-EM structure of yeast RNA polymerase III at 4.7 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5OA1
 
 | | RNA polymerase I pre-initiation complex | | Descriptor: | ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Sadian, Y, Tafur, L, Kosinski, J, Jakobi, A.J, Muller, C.W. | | Deposit date: | 2017-06-20 | | Release date: | 2017-07-26 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into transcription initiation by yeast RNA polymerase I.
EMBO J., 36, 2017
|
|
