7BEW
 
 | |
7BEX
 
 | |
2WGQ
 
 | |
2GHC
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
1MPJ
 
 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | CHLORIDE ION, PHENOL, PHENOL INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
7ZDQ
 
 | | Cryo-EM structure of Human ACE2 bound to a high-affinity SARS CoV-2 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Bate, N, Savva, C.G, Moody, P.C.E, Brown, E.A, Schwabe, W.R, Brindle, N.P.J, Ball, J.K, Sale, J.E. | | Deposit date: | 2022-03-29 | | Release date: | 2022-05-18 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding.
Plos Pathog., 18, 2022
|
|
1VYP
 
 | | Structure of pentaerythritol tetranitrate reductase W102F mutant and complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
6XV4
 
 | | Neutron structure of ferric ascorbate peroxidase-ascorbate complex | | Descriptor: | ASCORBIC ACID, Ascorbate peroxidase, POTASSIUM ION, ... | | Authors: | Kwon, H, Basran, J, Devos, J.M, Schrader, T.E, Ostermann, A, Blakeley, M.P, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2020-01-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the protons in a metalloenzyme electron proton transfer pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1VYR
 
 | | Structure of pentaerythritol tetranitrate reductase complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-05 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
2CLA
 
 | |
5LGZ
 
 | | Structure of Photoreduced Pentaerythritol Tetranitrate Reductase | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ISOPROPYL ALCOHOL, Pentaerythritol tetranitrate reductase | | Authors: | Kwon, H, Smith, O.M, Moody, P.C.E. | | Deposit date: | 2016-07-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combining X-ray and neutron crystallography with spectroscopy.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5LGX
 
 | |
6QPJ
 
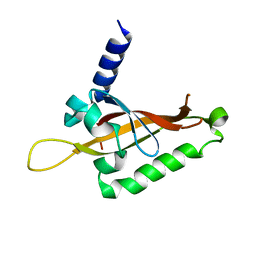 | | Human CLOCK PAS-A domain | | Descriptor: | Circadian locomoter output cycles protein kaput | | Authors: | Kwon, H, Freeman, S.L, Moody, P.C.E, Raven, E.L, Basran, J. | | Deposit date: | 2019-02-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | Heme binding to human CLOCK affects interactions with the E-box.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7P46
 
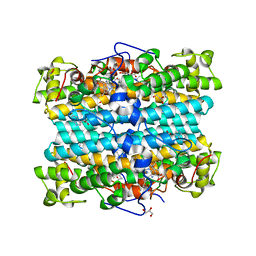 | | Crystal Structure of Xanthomonas campestris Tryptophan 2,3-dioxygenase (TDO) | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, CYANIDE ION, GLYCEROL, ... | | Authors: | Kwon, H, Basran, J, Booth, E.S, Campbell, L.P, Thackray, S.J, Moody, P.C.E, Mowat, C.G, Raven, E.L. | | Deposit date: | 2021-07-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of l-kynurenine to X. campestris tryptophan 2,3-dioxygenase.
J.Inorg.Biochem., 225, 2021
|
|
244D
 
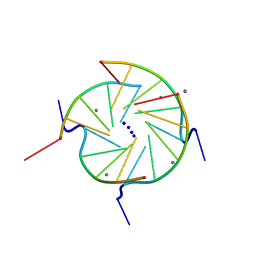 | | THE HIGH-RESOLUTION CRYSTAL STRUCTURE OF A PARALLEL-STRANDED GUANINE TETRAPLEX | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Laughlan, G, Murchie, A.I.H, Norman, D.G, Moore, M.H, Moody, P.C.E, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1995-10-19 | | Release date: | 1996-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The high-resolution crystal structure of a parallel-stranded guanine tetraplex.
Science, 265, 1994
|
|
1SFE
 
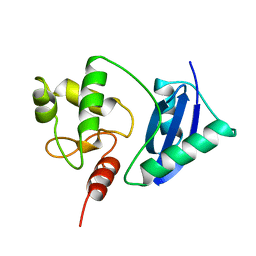 | | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE | | Authors: | Moore, M.H, Gulbis, J.M, Dodson, E.J, Demple, B, Moody, P.C.E. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli.
EMBO J., 13, 1994
|
|
5JPR
 
 | | Neutron Structure of Compound II of Ascorbate Peroxidase | | Descriptor: | Ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kwon, H, Blakeley, M.P, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2016-05-04 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.806 Å), X-RAY DIFFRACTION | | Cite: | Direct visualization of a Fe(IV)-OH intermediate in a heme enzyme.
Nat Commun, 7, 2016
|
|
5JQR
 
 | | The Structure of Ascorbate Peroxidase Compound II formed by reaction with m-CPBA | | Descriptor: | Ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kwon, H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2016-05-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Direct visualization of a Fe(IV)-OH intermediate in a heme enzyme.
Nat Commun, 7, 2016
|
|
1VYS
 
 | | STRUCTURE OF PENTAERYTHRITOL TETRANITRATE REDUCTASE W102Y MUTANT AND COMPLEXED WITH PICRIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-05 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
6TAE
 
 | | Neutron structure of ferric ascorbate peroxidase | | Descriptor: | Ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kwon, H, Basran, J, Devos, J.M, Schrader, T.E, Ostermann, A, Blakeley, M.P, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the protons in a metalloenzyme electron proton transfer pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2CL4
 
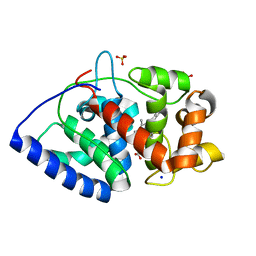 | | Ascorbate Peroxidase R172A mutant | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Ghamsari, L, MacDonald, I.K, Moody, P.C.E. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of Ascorbate Peroxidase with Substrates: A Mechanistic and Structural Analysis.
Biochemistry, 45, 2006
|
|
4LOR
 
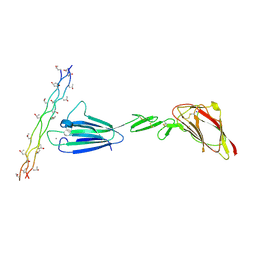 | | C1s CUB1-EGF-CUB2 in complex with a collagen-like peptide from C1q | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1s subcomponent heavy chain, ... | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LMF
 
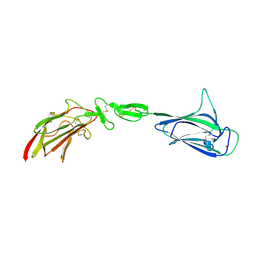 | | C1s CUB1-EGF-CUB2 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain, SODIUM ION | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.921 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LOS
 
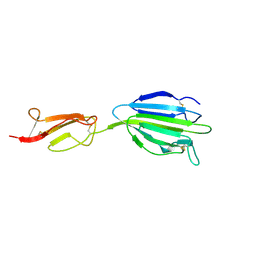 | | C1s CUB2-CCP1 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E, Gingras, A.R. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LOT
 
 | | C1s CUB2-CCP1-CCP2 | | Descriptor: | Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
