1P31
 
 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine Ligase (MurC) from Haemophilus influenzae | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--alanine ligase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-16 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
1P3D
 
 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine ligase (MurC) in Complex with UMA and ANP. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--alanine ligase, ... | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
1PKG
 
 | | Structure of a c-Kit Kinase Product Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, c-kit protein | | Authors: | Mol, C.D, Lim, K.B, Sridhar, V, Zou, H, Chien, E.Y.T, Sang, B.-C, Nowakowski, J, Kassel, D.B, Cronin, C.N, McRee, D.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a c-Kit Product Complex Reveals the Basis for Kinase Transactivation.
J.Biol.Chem., 278, 2003
|
|
1Q5H
 
 | | Human dUTP Pyrophosphatase complex with dUDP | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, dUTP pyrophosphatase | | Authors: | Mol, C.D, Harris, J.M, McIntosh, E.M, Tainer, J.A. | | Deposit date: | 2003-08-07 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human dUTP pyrophosphatase: uracil recognition by a Beta hairpin and active sites formed by three separate subunits
Structure, 4, 1996
|
|
1Q5U
 
 | | HUMAN DUTP PYROPHOSPHATASE | | Descriptor: | dUTP pyrophosphatase | | Authors: | Mol, C.D, Harris, J.M, Mcintosh, E.M, Tainer, J.A. | | Deposit date: | 2003-08-11 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human dUTP Pyrophosphatase: Uracil Recognition by a Beta Hairpin and Active Sites Formed by Three Separate Subunits
Structure, 4, 1996
|
|
1KEA
 
 | | STRUCTURE OF A THERMOSTABLE THYMINE-DNA GLYCOSYLASE | | Descriptor: | ACETATE ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Mol, C.D, Arvai, A.S, Begley, T.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 2001-11-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases.
J.Mol.Biol., 315, 2002
|
|
3GB2
 
 | | GSK3beta inhibitor complex | | Descriptor: | 2-methyl-5-(3-{4-[(S)-methylsulfinyl]phenyl}-1-benzofuran-5-yl)-1,3,4-oxadiazole, Glycogen synthase kinase-3 beta | | Authors: | Mol, C.D. | | Deposit date: | 2009-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-{3-[4-(Alkylsulfinyl)phenyl]-1-benzofuran-5-yl}-5-methyl-1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta with good brain permeability.
J.Med.Chem., 52, 2009
|
|
1UGH
 
 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE IN COMPLEX WITH A PROTEIN INHIBITOR: PROTEIN MIMICRY OF DNA | | Descriptor: | PROTEIN (URACIL-DNA GLYCOSYLASE INHIBITOR), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Mol, C.D, Arvai, A.S, Sanderson, R.J, Slupphaug, G, Kavli, B, Krokan, H.E, Mosbaugh, D.W, Tainer, J.A. | | Deposit date: | 1999-02-05 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA.
Cell(Cambridge,Mass.), 82, 1995
|
|
1AKO
 
 | | EXONUCLEASE III FROM ESCHERICHIA COLI | | Descriptor: | EXONUCLEASE III | | Authors: | Mol, C.D, Kuo, C.-F, Thayer, M.M, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the multifunctional DNA-repair enzyme exonuclease III.
Nature, 374, 1995
|
|
1UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
3F7Z
 
 | | X-ray Co-Crystal Structure of Glycogen Synthase Kinase 3beta in Complex with an Inhibitor | | Descriptor: | 2-(1,3-benzodioxol-5-yl)-5-[(3-fluoro-4-methoxybenzyl)sulfanyl]-1,3,4-oxadiazole, Glycogen synthase kinase-3 beta | | Authors: | Mol, C.D, Dougan, D.R. | | Deposit date: | 2008-11-10 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, synthesis and structure-activity relationships of 1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta.
Bioorg.Med.Chem., 17, 2009
|
|
3F88
 
 | | glycogen synthase Kinase 3beta inhibitor complex | | Descriptor: | 3-methylbenzonitrile, 5-[1-(4-methoxyphenyl)-1H-benzimidazol-6-yl]-1,3,4-oxadiazole-2(3H)-thione, Glycogen synthase kinase-3 beta | | Authors: | Mol, C.D, Dougan, D.R. | | Deposit date: | 2008-11-11 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and structure-activity relationships of 1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta.
Bioorg.Med.Chem., 17, 2009
|
|
1DE8
 
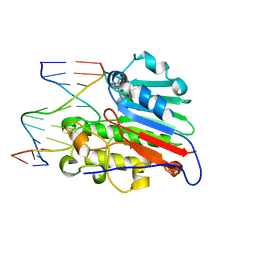 | | HUMAN APURINIC/APYRIMIDINIC ENDONUCLEASE-1 (APE1) BOUND TO ABASIC DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*CP*(3DR)P*GP*AP*TP*CP*G)-3'), MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected
Nature, 403, 2000
|
|
1DE9
 
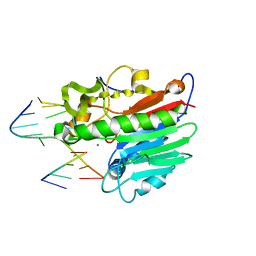 | | HUMAN APE1 ENDONUCLEASE WITH BOUND ABASIC DNA AND MN2+ ION | | Descriptor: | 5'-d(*CP*TP*AP*C)-3', 5'-d(*GP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-d(P*(3DR)P*GP*AP*TP*C)-3', ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
1DEW
 
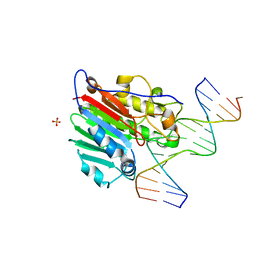 | | CRYSTAL STRUCTURE OF HUMAN APE1 BOUND TO ABASIC DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*(3DR)P*CP*GP*AP*CP*GP*AP*CP*G)-3', 5'-D(*GP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*C)-3', MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE, ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-15 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
1T46
 
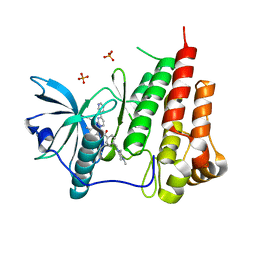 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, PHOSPHATE ION | | Authors: | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
1T45
 
 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | Descriptor: | Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | | Authors: | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
4SKN
 
 | | A NUCLEOTIDE-FLIPPING MECHANISM FROM THE STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*CP*GP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*(D1P)P*GP*GP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE), ... | | Authors: | Slupphaug, G, Mol, C.D, Kavli, B, Arvai, A.S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-02-20 | | Release date: | 1999-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA.
Nature, 384, 1996
|
|
2UGI
 
 | | PROTEIN MIMICRY OF DNA FROM CRYSTAL STRUCTURES OF THE URACIL GLYCOSYLASE INHIBITOR PROTEIN AND ITS COMPLEX WITH ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | Descriptor: | IMIDAZOLE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-11-06 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
2UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH H187D MUTANT UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
4IEM
 
 | | Human apurinic/apyrimidinic endonuclease (APE1) with product DNA and Mg2+ | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*C)-3'), DNA (5'-D(P*(3DR)P*GP*AP*TP*CP*G)-3'), ... | | Authors: | Tsutakawa, S.E, Mol, C.D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3936 Å) | | Cite: | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
4LGU
 
 | | Crystal structure of clAP1 BIR3 bound to T3226692 | | Descriptor: | (3S,8aR)-N-((R)-chroman-4-yl)-2-((S)-2-cyclohexyl-2-((S)-2-(methylamino)propanamido)acetyl)octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Mol, C.D, Snell, G.P. | | Deposit date: | 2013-06-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, stereoselective synthesis, and biological evaluation of novel tri-cyclic compounds as inhibitor of apoptosis proteins (IAP) antagonists.
Bioorg.Med.Chem., 21, 2013
|
|
3TDC
 
 | | Crystal Structure of Human Acetyl-CoA carboxylase 2 | | Descriptor: | 1-[3-({4-[(5S)-3,3-dimethyl-1-oxo-2-oxa-7-azaspiro[4.5]dec-7-yl]piperidin-1-yl}carbonyl)-1-benzothiophen-2-yl]-3-ethylurea, Acetyl-CoA carboxylase 2 variant | | Authors: | Dougan, D.R, Mol, C.D. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Design, synthesis, and structure-activity relationships of spirolactones bearing 2-ureidobenzothiophene as acetyl-CoA carboxylases inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OE0
 
 | | Crystal structure of the CXCR4 chemokine receptor in complex with a cyclic peptide antagonist CVX15 | | Descriptor: | C-X-C chemokine receptor type 4, Lysozyme Chimera, Polyphemusin analog, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE9
 
 | | Crystal structure of the chemokine CXCR4 receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
